Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts
- PMID: 19386601
- PMCID: PMC2719312
- DOI: 10.1074/jbc.M901790200
Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts
Abstract
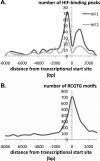
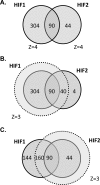
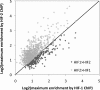
Hypoxia-inducible factor (HIF) controls an extensive range of adaptive responses to hypoxia. To better understand this transcriptional cascade we performed genome-wide chromatin immunoprecipitation using antibodies to two major HIF-alpha subunits, and correlated the results with genome-wide transcript profiling. Within a tiled promoter array we identified 546 and 143 sequences that bound, respectively, to HIF-1alpha or HIF-2alpha at high stringency. Analysis of these sequences confirmed an identical core binding motif for HIF-1alpha and HIF-2alpha (RCGTG) but demonstrated that binding to this motif was highly selective, with binding enriched at distinct regions both upstream and downstream of the transcriptional start. Comparison of HIF-promoter binding data with bidirectional HIF-dependent changes in transcript expression indicated that whereas a substantial proportion of positive responses (>20% across all significantly regulated genes) are direct, HIF-dependent gene suppression is almost entirely indirect. Comparison of HIF-1alpha- versus HIF-2alpha-binding sites revealed that whereas some loci bound HIF-1alpha in isolation, many bound both isoforms with similar affinity. Despite high-affinity binding to multiple promoters, HIF-2alpha contributed to few, if any, of the transcriptional responses to acute hypoxia at these loci. Given emerging evidence for biologically distinct functions of HIF-1alpha versus HIF-2alpha understanding the mechanisms restricting HIF-2alpha activity will be of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

