Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley
- PMID: 19386806
- PMCID: PMC2689963
- DOI: 10.1104/pp.109.137901
Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley
Abstract
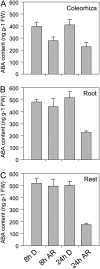
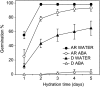
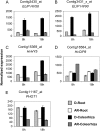
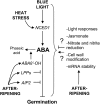
The decay of seed dormancy during after-ripening is not well understood, but elucidation of the mechanisms involved may be important for developing strategies for modifying dormancy in crop species and, for example, addressing the problem of preharvest sprouting in cereals. We have studied the germination characteristics of barley (Hordeum vulgare 'Betzes') embryos, including a description of anatomical changes in the coleorhiza and the enclosed seminal roots. The changes that occur correlate with abscisic acid (ABA) contents of embryo tissues. To understand the molecular mechanisms involved in dormancy loss, we compared the transcriptome of dormant and after-ripened barley embryos using a tissue-specific microarray approach. Our results indicate that in the coleorhiza, ABA catabolism is promoted and ABA sensitivity is reduced and that this is associated with differential regulation by after-ripening of ABA 8'-hydroxylase and of the LIPID PHOSPHATE PHOSPHATASE gene family and ABI3-INTERACTING PROTEIN2, respectively. We also identified other processes, including jasmonate responses, cell wall modification, nitrate and nitrite reduction, mRNA stability, and blue light sensitivity, that were affected by after-ripening in the coleorhiza that may be downstream of ABA signaling. Based on these results, we propose that the coleorhiza plays a major role in causing dormancy by acting as a barrier to root emergence and that after-ripening potentiates molecular changes related to ABA metabolism and sensitivity that ultimately lead to degradation of the coleorhiza, root emergence, and germination.
Figures






References
-
- Avery GS (1930) Comparative anatomy and morphology of embryos and seedlings of maize, oats and wheat. Bot Gaz 89 1–39
-
- Barrero JM, Jacobsen J, Gubler F (2009) Seed dormancy: approaches for finding new genes in cereals. In E-C Pua, M Davey, eds, Plant Developmental Biology: Biotechnological Perspectives. Springer, Berlin (in press)
-
- Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29 2000–2008 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

