Filaggrin in the frontline: role in skin barrier function and disease
- PMID: 19386895
- PMCID: PMC2721001
- DOI: 10.1242/jcs.033969
Filaggrin in the frontline: role in skin barrier function and disease
Abstract
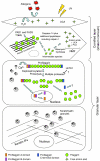
Recently, loss-of-function mutations in FLG, the human gene encoding profilaggrin and filaggrin, have been identified as the cause of the common skin condition ichthyosis vulgaris (which is characterised by dry, scaly skin). These mutations, which are carried by up to 10% of people, also represent a strong genetic predisposing factor for atopic eczema, asthma and allergies. Profilaggrin is the major component of the keratohyalin granules within epidermal granular cells. During epidermal terminal differentiation, the approximately 400 kDa profilaggrin polyprotein is dephosphorylated and rapidly cleaved by serine proteases to form monomeric filaggrin (37 kDa), which binds to and condenses the keratin cytoskeleton and thereby contributes to the cell compaction process that is required for squame biogenesis. Within the squames, filaggrin is citrullinated, which promotes its unfolding and further degradation into hygroscopic amino acids, which constitute one element of natural moisturising factor. Loss of profilaggrin or filaggrin leads to a poorly formed stratum corneum (ichthyosis), which is also prone to water loss (xerosis). Recent human genetic studies strongly suggest that perturbation of skin barrier function as a result of reduction or complete loss of filaggrin expression leads to enhanced percutaneous transfer of allergens. Filaggrin is therefore in the frontline of defence, and protects the body from the entry of foreign environmental substances that can otherwise trigger aberrant immune responses.
Figures




References
-
- Brown, S. J., Relton, C. L., Liao, H., Zhao, Y., Sandilands, A., Wilson, I. J., Burn, J., Reynolds, N. J., McLean, W. H. I. and Cordell, H. J. (2008). Filaggrin null mutations and childhood atopic eczema: a population-based case-control study. J. Allergy Clin. Immunol. 121, 940-946.e3. - PMC - PubMed
-
- Chavanas, S., Bodemer, C., Rochat, A., Hamel-Teillac, D., Ali, M., Irvine, A. D., Bonafe, J. L., Wilkinson, J., Taieb, A., Barrandon, Y. et al. (2000). Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 25, 141-142. - PubMed
-
- Chen, H., Ho, J. C., Sandilands, A., Chan, Y. C., Giam, Y. C., Evans, A. T., Lane, E. B. and McLean, W. H. I. (2008). Unique and recurrent mutations in the filaggrin gene in Singaporean Chinese patients with ichthyosis vulgaris. J. Invest. Dermatol. 128, 1669-1675. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

