Dinoflagellate spliced leader RNA genes display a variety of sequences and genomic arrangements
- PMID: 19387009
- PMCID: PMC2734150
- DOI: 10.1093/molbev/msp083
Dinoflagellate spliced leader RNA genes display a variety of sequences and genomic arrangements
Abstract
Spliced leader (SL) trans-splicing is a common mRNA processing mechanism in dinoflagellates, in which a 22-nt sequence is transferred from the 5'-end of a small noncoding RNA, the SL RNA, to the 5'-end of mRNA molecules. Although the SL RNA gene was shown initially to be organized as tandem repeats with transcripts of 50-60 nt, shorter than most of their counterparts in other organisms, other gene organizations and transcript lengths were reported subsequently. To address the evolutionary gradient of gene organization complexity, we thoroughly examined transcript and gene organization of the SL RNA in a phylogenetically and ecologically diverse group of dinoflagellates representing four Orders. All these dinoflagellates possessed SL RNA transcripts of 50-60 nt, although in one species additional transcripts of up to 92 nt were also detected. At the genomic level, various combinations of SL RNA and 5S rRNA tandem gene arrays, including SL RNA-only, 5S rRNA-only, and mixed SL RNA-5S rRNA (SL-5S) clusters, were amplified by polymerase chain reaction for six dinoflagellates, containing intergenic spacers ranging from 88 bp to over 1.2 kb. Of these species, no SL-5S cluster was detected in Prorocentrum minimum, and only Karenia brevis showed the U6 small nuclear RNA gene associated with these mixed arrays. The 5S rRNA-only array was also found in three dinoflagellates, along with two SL-5S-adjacent arrangements found in two other species that could represent junctions. Two species contained multimeric SL exon repeats with no associated intron. These results suggest that 1) both the SL RNA tandem repeat and the SL-5S cluster genomic organizations are an "ancient" and widespread feature within the phylum of dinoflagellates and 2) rampant genomic duplication and recombination are ongoing independently in each dinoflagellate lineage, giving rise to the highly complex and diversified genomic arrangements of the SL RNA gene, while conserving the length and structure of the functional SL RNA.
Figures
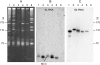



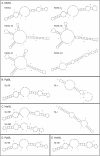
References
-
- Blaxter ML, Liu LX. Nematode spliced leaders: ubiquity, evolution and utility. Int J Parasitol. 1996;26:1025–1033. - PubMed
-
- Blumenthal T. Tran-splicing and operons. In: WormBook, editor. The C. elegans Research Community, WormBook. 2005. doi/10.1895/wormbook.1.5.1 http://www.wormbook.org. - PubMed
-
- Boothroyd JC, Cross GAM. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. - PubMed
-
- Bruzik JP, Doren KV, Hirsh D, Steitz JA. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

