Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics
- PMID: 19387468
- PMCID: PMC2950785
- DOI: 10.1038/leu.2009.82
Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics
Abstract
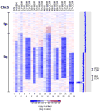
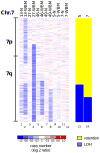
Progress in the management of patients with myelodysplastic syndromes (MDS) has been hampered by the inability to detect cytogenetic abnormalities in 40-60% of cases. We prospectively analyzed matched pairs of bone marrow and buccal cell (normal) DNA samples from 51 MDS patients by single nucleotide polymorphism (SNP) arrays, and identified somatically acquired clonal genomic abnormalities in 21 patients (41%). Among the 33 patients with normal bone marrow cell karyotypes, 5 (15%) had clonal, somatically acquired aberrations by SNP array analysis, including 4 with segmental uniparental disomies (UPD) and 1 with three separate microdeletions. Each abnormality was detected more readily in CD34+ cells than in unselected bone marrow cells. Paired analysis of bone marrow and buccal cell DNA from each patient was necessary to distinguish true clonal genomic abnormalities from inherited copy number variations and regions with apparent loss of heterozygosity. UPDs affecting chromosome 7q were identified in two patients who had a rapidly deteriorating clinical course despite a low-risk International Prognostic Scoring System score. Further studies of larger numbers of patients will be needed to determine whether 7q UPD detected by SNP array analysis will identify higher risk MDS patients at diagnosis, analogous to those with 7q cytogenetic abnormalities.
Figures




References
-
- Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007 Feb;7(2):118–129. - PubMed
-
- Estey E. Acute myeloid leukemia and myelodysplastic syndromes in older patients. J Clin Oncol. 2007 May 10;25(14):1908–1915. - PubMed
-
- Nimer SD. Myelodysplastic syndromes. Blood. 2008 May 15;111(10):4841–4851. - PubMed
-
- de Witte T, Oosterveld M, Muus P. Autologous and allogeneic stem cell transplantation for myelodysplastic syndrome. Blood Rev. 2007 Jan;21(1):49–59. - PubMed
-
- Haase D, Germing U, Schanz J, Pfeilstocker M, Nosslinger T, Hildebrandt B, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007 Dec 15;110(13):4385–4395. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

