S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains
- PMID: 19387488
- PMCID: PMC2711177
- DOI: 10.1038/emboj.2009.115
S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains
Abstract
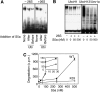
Ubiquitin (Ub)-protein conjugates formed by purified ring-finger or U-box E3s with the E2, UbcH5, resist degradation and disassembly by 26S proteasomes. These chains contain multiple types of Ub forks in which two Ub's are linked to adjacent lysines on the proximal Ub. We tested whether cells contain factors that prevent formation of nondegradable conjugates and whether the forked chains prevent proteasomal degradation. S5a is a ubiquitin interacting motif (UIM) protein present in the cytosol and in the 26S proteasome. Addition of S5a or a GST-fusion of S5a's UIM domains to a ubiquitination reaction containing 26S proteasomes, UbcH5, an E3 (MuRF1 or CHIP), and a protein substrate, dramatically stimulated its degradation, provided S5a was present during ubiquitination. Mass spectrometry showed that S5a and GST-UIM prevented the formation of Ub forks without affecting synthesis of standard isopeptide linkages. The forked Ub chains bind poorly to 26S proteasomes unlike those synthesized with S5a present or linked to Lys63 or Lys48 chains. Thus, S5a (and presumably certain other UIM proteins) function with certain E3/E2 pairs to ensure synthesis of efficiently degraded non-forked Ub conjugates.
Figures


Comment in
-
Nonconformity in ubiquitin compliance.EMBO J. 2009 Jul 8;28(13):1825-7. doi: 10.1038/emboj.2009.132. EMBO J. 2009. PMID: 19587678 Free PMC article. No abstract available.
References
-
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 - PubMed
-
- Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40: 427–446 - PubMed
-
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

