Tryptophan synthase: a mine for enzymologists
- PMID: 19387555
- PMCID: PMC11115766
- DOI: 10.1007/s00018-009-0028-0
Tryptophan synthase: a mine for enzymologists
Abstract
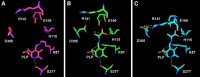
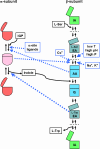
Tryptophan synthase is a pyridoxal 5'-phosphate-dependent alpha(2)beta(2) complex catalyzing the last two steps of tryptophan biosynthesis in bacteria, plants and fungi. Structural, dynamic and functional studies, carried out over more than 40 years, have unveiled that: (1) alpha- and beta-active sites are separated by about 20 A and communicate via the selective stabilization of distinct conformational states, triggered by the chemical nature of individual catalytic intermediates and by allosteric ligands; (2) indole, formed at alpha-active site, is intramolecularly channeled to the beta-active site; and (3) naturally occurring as well as genetically generated mutants have allowed to pinpoint functional and regulatory roles for several individual amino acids. These key features have made tryptophan synthase a text-book case for the understanding of the interplay between chemistry and conformational energy landscapes.
Figures


Similar articles
-
On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.J Mol Biol. 2002 Dec 6;324(4):677-90. doi: 10.1016/s0022-2836(02)01109-9. J Mol Biol. 2002. PMID: 12460570
-
Catalytically impaired TrpA subunit of tryptophan synthase from Chlamydia trachomatis is an allosteric regulator of TrpB.Protein Sci. 2021 Sep;30(9):1904-1918. doi: 10.1002/pro.4143. Epub 2021 Jun 16. Protein Sci. 2021. PMID: 34107106 Free PMC article.
-
Tryptophan synthase, an allosteric molecular factory.Curr Opin Chem Biol. 2008 Oct;12(5):593-600. doi: 10.1016/j.cbpa.2008.07.011. Curr Opin Chem Biol. 2008. PMID: 18675375
-
Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex.Arch Biochem Biophys. 2012 Mar 15;519(2):154-66. doi: 10.1016/j.abb.2012.01.016. Epub 2012 Feb 2. Arch Biochem Biophys. 2012. PMID: 22310642 Free PMC article. Review.
-
Tryptophan synthase: a multienzyme complex with an intramolecular tunnel.Chem Rec. 2001;1(2):140-51. doi: 10.1002/tcr.4. Chem Rec. 2001. PMID: 11893063 Review.
Cited by
-
Plant growth-promoting properties of Streptomyces spp. isolates and their impact on mung bean plantlets' rhizosphere microbiome.Front Microbiol. 2022 Aug 25;13:967415. doi: 10.3389/fmicb.2022.967415. eCollection 2022. Front Microbiol. 2022. PMID: 36090067 Free PMC article.
-
The role of oligomerization and cooperative regulation in protein function: the case of tryptophan synthase.PLoS Comput Biol. 2010 Nov 11;6(11):e1000994. doi: 10.1371/journal.pcbi.1000994. PLoS Comput Biol. 2010. PMID: 21085641 Free PMC article.
-
Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.Biochemistry. 2013 Sep 17;52(37):6396-411. doi: 10.1021/bi400795e. Epub 2013 Sep 6. Biochemistry. 2013. PMID: 23952479 Free PMC article.
-
Additive and Emergent Catalytic Properties of Dimeric Unnatural Amino Acid Derivatives: Aldol and Conjugate Additions.Chemistry. 2021 Nov 11;27(63):15671-15687. doi: 10.1002/chem.202102394. Epub 2021 Oct 13. Chemistry. 2021. PMID: 34453455 Free PMC article.
-
Allosteric regulation of substrate channeling: Salmonella typhimurium tryptophan synthase.Front Mol Biosci. 2022 Sep 12;9:923042. doi: 10.3389/fmolb.2022.923042. eCollection 2022. Front Mol Biosci. 2022. PMID: 36172042 Free PMC article. Review.
References
-
- Barends TR, Dunn MF, Schlichting I. Tryptophan synthase, an allosteric molecular factory. Curr Opin Chem Biol. 2008;12:593–600. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

