Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages
- PMID: 19388904
- PMCID: PMC2746821
- DOI: 10.1111/j.1462-5822.2009.01316.x
Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages
Abstract
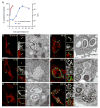
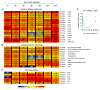
Summary The highly infectious bacterium Francisella tularensis is a facultative intracellular pathogen, whose virulence requires proliferation inside host cells, including macrophages. Here we have performed a global transcriptional profiling of the highly virulent F. tularensis ssp. tularensis Schu S4 strain during its intracellular cycle within primary murine macrophages, to characterize its intracellular biology and identify pathogenic determinants based on their intracellular expression profiles. Phagocytosed bacteria rapidly responded to their intracellular environment and subsequently altered their transcriptional profile. Differential gene expression profiles were revealed that correlated with specific intracellular locale of the bacteria. Upregulation of general and oxidative stress response genes was a hallmark of the early phagosomal and late endosomal stages, while induction of transport and metabolic genes characterized the cytosolic replication stage. Expression of the Francisella Pathogenicity Island (FPI) genes, which are required for intracellular proliferation, increased during the intracellular cycle. Similarly, 27 chromosomal loci encoding putative hypothetical, secreted, outer membrane proteins or transcriptional regulators were identified as upregulated. Among these, deletion of FTT0383, FTT0369c or FTT1676 abolished the ability of Schu S4 to survive or proliferate intracellularly and cause lethality in mice, therefore identifying novel determinants of Francisella virulence from their intracellular expression profile.
Figures



Similar articles
-
The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression.Infect Immun. 2008 Dec;76(12):5488-99. doi: 10.1128/IAI.00682-08. Epub 2008 Oct 13. Infect Immun. 2008. PMID: 18852245 Free PMC article.
-
Whole genome transcriptomics reveals global effects including up-regulation of Francisella pathogenicity island gene expression during active stringent response in the highly virulent Francisella tularensis subsp. tularensis SCHU S4.Microbiology (Reading). 2017 Nov;163(11):1664-1679. doi: 10.1099/mic.0.000550. Epub 2017 Oct 16. Microbiology (Reading). 2017. PMID: 29034854 Free PMC article.
-
Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis.Infect Immun. 2009 May;77(5):1866-80. doi: 10.1128/IAI.01496-08. Epub 2009 Feb 17. Infect Immun. 2009. PMID: 19223477 Free PMC article.
-
Loops and networks in control of Francisella tularensis virulence.Future Microbiol. 2009 Aug;4(6):713-29. doi: 10.2217/fmb.09.37. Future Microbiol. 2009. PMID: 19659427 Review.
-
Proteins involved in Francisella tularensis survival and replication inside macrophages.Future Microbiol. 2012 Nov;7(11):1255-68. doi: 10.2217/fmb.12.103. Future Microbiol. 2012. PMID: 23075445 Review.
Cited by
-
Immune lymphocytes halt replication of Francisella tularensis LVS within the cytoplasm of infected macrophages.Sci Rep. 2020 Jul 21;10(1):12023. doi: 10.1038/s41598-020-68798-2. Sci Rep. 2020. PMID: 32694562 Free PMC article.
-
Francisella philomiragia Infection and Lethality in Mammalian Tissue Culture Cell Models, Galleria mellonella, and BALB/c Mice.Front Microbiol. 2016 May 24;7:696. doi: 10.3389/fmicb.2016.00696. eCollection 2016. Front Microbiol. 2016. PMID: 27252681 Free PMC article.
-
Comparative transcriptional analysis of homologous pathogenic and non-pathogenic Lawsonia intracellularis isolates in infected porcine cells.PLoS One. 2012;7(10):e46708. doi: 10.1371/journal.pone.0046708. Epub 2012 Oct 3. PLoS One. 2012. PMID: 23056413 Free PMC article.
-
Critical Role of a Sheath Phosphorylation Site On the Assembly and Function of an Atypical Type VI Secretion System.Mol Cell Proteomics. 2019 Dec;18(12):2418-2432. doi: 10.1074/mcp.RA119.001532. Epub 2019 Oct 2. Mol Cell Proteomics. 2019. PMID: 31578219 Free PMC article.
-
The reduced genome of the Francisella tularensis live vaccine strain (LVS) encodes two iron acquisition systems essential for optimal growth and virulence.PLoS One. 2014 Apr 2;9(4):e93558. doi: 10.1371/journal.pone.0093558. eCollection 2014. PLoS One. 2014. PMID: 24695402 Free PMC article.
References
-
- Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, et al. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Molecular microbiology. 2003;50:219–230. - PubMed
-
- Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Molecular microbiology. 1998;29:247–259. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

