Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes
- PMID: 19390088
- PMCID: PMC2675864
- DOI: 10.1101/gad.524609
Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes
Abstract
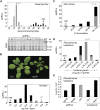
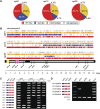
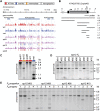
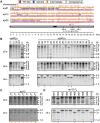
Transgenerational epigenetic inheritance has been defined by the study of relatively few loci. We examined a population of recombinant inbred lines with epigenetically mosaic chromosomes consisting of wild-type and CG methylation-depleted segments (epiRILs). Surprisingly, transposons that were immobile in the parental lines displayed stochastic movement in 28% of the epiRILs. Although analysis after eight generations of inbreeding, supported by genome-wide DNA methylation profiling, identified recombined parental chromosomal segments, these were interspersed with unexpectedly high frequencies of nonparental methylation polymorphism. Hence, epigenetic inheritance in hybrids derived from parents with divergent epigenomes permits long-lasting epi-allelic interactions that violate Mendelian expectations. Such persistently "unstable" epigenetic states complicate linkage-based epigenomic mapping. Thus, future epigenomic analyses should consider possible genetic instabilities and alternative mapping strategies.
Figures



References
-
- Bao N., Lye K.W., Barton M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:653–662. - PubMed
-
- Chan S.W., Henderson I.R., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. - PubMed
-
- Fan J., Crooks C., Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53:393–399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
