A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis
- PMID: 19390092
- PMCID: PMC2675869
- DOI: 10.1101/gad.1781709
A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis
Abstract
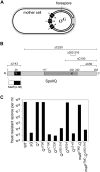
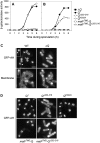
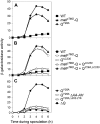
Spore formation by Bacillus subtilis takes place in a sporangium consisting of two chambers, the forespore and the mother cell, which are linked by pathways of intercellular communication. One pathway, which couples the activation of the forespore transcription factor sigma(G) to the action of sigma(E) in the mother cell, has remained mysterious. Traditional models hold that sigma(E) initiates a signal transduction pathway that specifically activates sigma(G) in the forespore. Recent experiments indicating that the mother cell and forespore are joined by a channel have led to the suggestion that a specific regulator of sigma(G) is transported from the mother cell into the forespore. As we report here, however, the requirement for the channel is not limited to sigma(G). Rather, it is also required for the persistent activity of the early-acting forespore transcription factor sigma(F) as well as that of a heterologous RNA polymerase (that of phage T7). We infer that macromolecular synthesis in the forespore becomes dependent on the channel at intermediate stages of development. We propose that the channel is a gap junction-like feeding tube through which the mother cell nurtures the developing spore by providing small molecules needed for biosynthetic activity, including sigma(G)-directed gene activation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
