Biological process linkage networks
- PMID: 19390589
- PMCID: PMC2669181
- DOI: 10.1371/journal.pone.0005313
Biological process linkage networks
Abstract
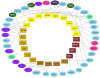
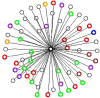
Background: The traditional approach to studying complex biological networks is based on the identification of interactions between internal components of signaling or metabolic pathways. By comparison, little is known about interactions between higher order biological systems, such as biological pathways and processes. We propose a methodology for gleaning patterns of interactions between biological processes by analyzing protein-protein interactions, transcriptional co-expression and genetic interactions. At the heart of the methodology are the concept of Linked Processes and the resultant network of biological processes, the Process Linkage Network (PLN).
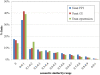
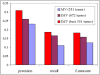
Results: We construct, catalogue, and analyze different types of PLNs derived from different data sources and different species. When applied to the Gene Ontology, many of the resulting links connect processes that are distant from each other in the hierarchy, even though the connection makes eminent sense biologically. Some others, however, carry an element of surprise and may reflect mechanisms that are unique to the organism under investigation. In this aspect our method complements the link structure between processes inherent in the Gene Ontology, which by its very nature is species-independent. As a practical application of the linkage of processes we demonstrate that it can be effectively used in protein function prediction, having the power to increase both the coverage and the accuracy of predictions, when carefully integrated into prediction methods.
Conclusions: Our approach constitutes a promising new direction towards understanding the higher levels of organization of the cell as a system which should help current efforts to re-engineer ontologies and improve our ability to predict which proteins are involved in specific biological processes.
Conflict of interest statement
Figures
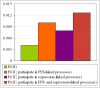
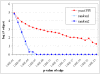
References
-
- Battle A, Segal E, Koller D. Probabilistic discovery of overlapping cellular processes and their regulation. J Comput Biol. 2005;12:909–927. - PubMed
-
- Bork P, et al. Protein interaction networks from yeast to human. Curr Opin Struct Biol. 2004;14:292–999. - PubMed
-
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;30:631–636. - PubMed
-
- Huynen MA, Snel B, von Mering C, Bork P. Function prediction and protein networks. Curr Opin Cell Biol. 2003;15:191–198. - PubMed
-
- de Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

