Common gene variants in the tumor necrosis factor (TNF) and TNF receptor superfamilies and NF-kB transcription factors and non-Hodgkin lymphoma risk
- PMID: 19390683
- PMCID: PMC2669130
- DOI: 10.1371/journal.pone.0005360
Common gene variants in the tumor necrosis factor (TNF) and TNF receptor superfamilies and NF-kB transcription factors and non-Hodgkin lymphoma risk
Abstract
Background: A promoter polymorphism in the pro-inflammatory cytokine tumor necrosis factor (TNF) (TNF G-308A) is associated with increased non-Hodgkin lymphoma (NHL) risk. The protein product, TNF-alpha, activates the nuclear factor kappa beta (NF-kappaB) transcription factor, and is critical for inflammatory and apoptotic responses in cancer progression. We hypothesized that the TNF and NF-kappaB pathways are important for NHL and that gene variations across the pathways may alter NHL risk.
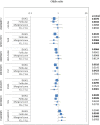
Methodology/principal findings: We genotyped 500 tag single nucleotide polymorphisms (SNPs) from 48 candidate gene regions (defined as 20 kb 5', 10 kb 3') in the TNF and TNF receptor superfamilies and the NF-kappaB and related transcription factors, in 1946 NHL cases and 1808 controls pooled from three independent population-based case-control studies. We obtained a gene region-level summary of association by computing the minimum p-value ("minP test"). We used logistic regression to compute odds ratios and 95% confidence intervals for NHL and four major NHL subtypes in relation to SNP genotypes and haplotypes. For NHL, the tail strength statistic supported an overall relationship between the TNF/NF-kappaB pathway and NHL (p = 0.02). We confirmed the association between TNF/LTA on chromosome 6p21.3 with NHL and found the LTA rs2844484 SNP most significantly and specifically associated with the major subtype, diffuse large B-cell lymphoma (DLBCL) (p-trend = 0.001). We also implicated for the first time, variants in NFKBIL1 on chromosome 6p21.3, associated with NHL. Other gene regions identified as statistically significantly associated with NHL included FAS, IRF4, TNFSF13B, TANK, TNFSF7 and TNFRSF13C. Accordingly, the single most significant SNPs associated with NHL were FAS rs4934436 (p-trend = 0.0024), IRF4 rs12211228 (p-trend = 0.0026), TNFSF13B rs2582869 (p-trend = 0.0055), TANK rs1921310 (p-trend = 0.0025), TNFSF7 rs16994592 (p-trend = 0.0024), and TNFRSF13C rs6002551 (p-trend = 0.0074). All associations were consistent in each study with no apparent specificity for NHL subtype.
Conclusions/significance: Our results provide consistent evidence that variation in the TNF superfamily of genes and specifically within chromosome 6p21.3 impacts lymphomagenesis. Further characterization of these susceptibility loci and identification of functional variants are warranted.
Conflict of interest statement
Figures

References
-
- Rothman N, Skibola CF, Wang SS, Morgan G, Lan Q, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006;1:27–38. - PubMed
-
- Wang SS, Cerhan JR, Hartge P, Davis S, Cozen W, et al. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-Hodgkin lymphoma. Cancer Res. 2006;19:9771–80. - PubMed
-
- Purdue MP, Lan Q, Kricker A, Grulich AE, Vajdic CM, et al. Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin Lymphoma Study. Carcinogenesis. 2007;3:704–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

