Prediction of the rotational tumbling time for proteins with disordered segments
- PMID: 19391622
- PMCID: PMC2694746
- DOI: 10.1021/ja809687r
Prediction of the rotational tumbling time for proteins with disordered segments
Abstract
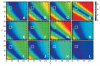
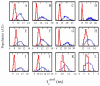
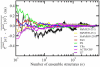
For well-structured, rigid proteins, the prediction of rotational tumbling time (tau(c)) using atomic coordinates is reasonably accurate, but is inaccurate for proteins with long unstructured sequences. Under physiological conditions, many proteins contain long disordered segments that play important regulatory roles in fundamental biological events including signal transduction and molecular recognition. Here we describe an ensemble approach to the boundary element method that accurately predicts tau(c) for such proteins by introducing two layers of molecular surfaces whose correlated velocities decay exponentially with distance. Reliable prediction of tau(c) will help to detect intra- and intermolecular interactions and conformational switches between more ordered and less ordered states of the disordered segments. The method has been extensively validated using 12 reference proteins with 14 to 103 disordered residues at the N- and/or C-terminus and has been successfully employed to explain a set of published results on a system that incorporates a conformational switch.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

