A whole-genome assembly of the domestic cow, Bos taurus
- PMID: 19393038
- PMCID: PMC2688933
- DOI: 10.1186/gb-2009-10-4-r42
A whole-genome assembly of the domestic cow, Bos taurus
Abstract
Background: The genome of the domestic cow, Bos taurus, was sequenced using a mixture of hierarchical and whole-genome shotgun sequencing methods.
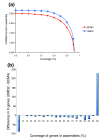
Results: We have assembled the 35 million sequence reads and applied a variety of assembly improvement techniques, creating an assembly of 2.86 billion base pairs that has multiple improvements over previous assemblies: it is more complete, covering more of the genome; thousands of gaps have been closed; many erroneous inversions, deletions, and translocations have been corrected; and thousands of single-nucleotide errors have been corrected. Our evaluation using independent metrics demonstrates that the resulting assembly is substantially more accurate and complete than alternative versions.
Conclusions: By using independent mapping data and conserved synteny between the cow and human genomes, we were able to construct an assembly with excellent large-scale contiguity in which a large majority (approximately 91%) of the genome has been placed onto the 30 B. taurus chromosomes. We constructed a new cow-human synteny map that expands upon previous maps. We also identified for the first time a portion of the B. taurus Y chromosome.
Figures




References
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. - DOI - PubMed
-
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, Hodgson A, George RA, Hoskins RA, Laverty T, Muzny DM, Nelson CR, Pacleb JM, Park S, Pfeiffer BD, Richards S, Sodergren EJ, Svirskas R, Tabor PE, Wan K, Stapleton M, Sutton GG, Venter C, Weinstock G, Scherer SE, Myers EW, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. - DOI - PMC - PubMed
-
- Snelling WM, Chiu R, Schein JE, Hobbs M, Abbey CA, Adelson DL, Aerts J, Bennett GL, Bosdet IE, Boussaha M, Brauning R, Caetano AR, Costa MM, Crawford AM, Dalrymple BP, Eggen A, Wind A Everts-van der, Floriot S, Gautier M, Gill CA, Green RD, Holt R, Jann O, Jones SJ, Kappes SM, Keele JW, de Jong PJ, Larkin DM, Lewin HA, McEwan JC, et al. A physical map of the bovine genome. Genome Biol. 2007;8:R165. doi: 10.1186/gb-2007-8-8-r165. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

