Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions
- PMID: 19393772
- PMCID: PMC2844463
- DOI: 10.1016/j.meegid.2009.04.014
Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions
Abstract
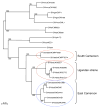
To evaluate the risk of cross-species transmissions of SIVs from non-human primates to humans at the primate/hunter interface, a total of 2586 samples, derived from primate bushmeat representing 11 different primate species, were collected at 6 distinct remote forest sites in southeastern Cameroon and in Yaoundé, the capital city. SIV prevalences were estimated with an updated SIV lineage specific gp41 peptide ELISA covering the major part of the SIV diversity. SIV positive samples were confirmed by PCR and sequence analysis of partial pol fragments. The updated SIV ELISA showed good performance with overall sensitivity and specificity of 96% and 97.5% respectively. The overall SIV seroprevalence was low, 2.93% (76/2586) and ranged between 0.0% and 5.7% at forest sites, and reached up to 10.3% in Yaoundé. SIV infection was documented in 8 of the 11 species with significantly different prevalence rates per species: 9/859 (1.0%) in Cercopithecus nictitans, 9/864 (1.0%) Cercopithecus cephus, 10/60 (16.7%) Miopithecus ogouensis, 14/78 (17.9%) Colobus guereza, 15/37 (40.5%) Cercopithecus neglectus, 10/27 (33.3%) Mandrillus sphinx, 6/12 (50%) Cercocebus torquatus, and 3/6 (50%) Chlorocebus tantalus. No SIV infection was identified in Cercopithecus pogonias (n=293), Lophocebus albigena (n=168) and Cercocebus agilis (n=182). The SIV prevalences also seem to vary within species according to the sampling site, but most importantly, the highest SIV prevalences are observed in the primate species which represent only 8.5% of the overall primate bushmeat. The phylogenetic tree of partial pol sequences illustrates the high genetic diversity of SIVs between and within different primate species. The tree also showed some interesting features within the SIVdeb lineage suggesting phylogeographic clusters. Overall, the risk for additional cross-species transmissions is not equal throughout southern Cameroon and depends on the hunted species and SIV prevalences in each species. However, humans are still exposed to a high diversity of SIVs as illustrated by the high inter and intra SIV lineage genetic diversity.
2009 Elsevier B.V. All rights reserved.
Figures

References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5. - PubMed
-
- Aghokeng AF, Bailes E, Loul S, Courgnaud V, Mpoudi-Ngolle E, Sharp PM, Delaporte E, Peeters M. Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages. Virology. 2007;360:407–18. - PMC - PubMed
-
- Aghokeng AF, Liu W, Bibollet-Ruche F, Loul S, Mpoudi-Ngole E, Laurent C, Mwenda JM, Langat DK, Chege GK, McClure HM, Delaporte E, Shaw GM, Hahn BH, Peeters M. Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology. 2006;345:174–89. - PubMed
-
- Auzel P, Hardin R. Colonial history, concessionary politics, and collaborative management of Equatorial African rain forests. In: Bakarr M, Da Fonseca G, Konstant W, Mittermeier R, Painemilla K, editors. Hunting and bushmeat utilization in the African rain forest. 2000. pp. 21–38.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

