Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex
- PMID: 19394292
- PMCID: PMC2727917
- DOI: 10.1016/j.molcel.2009.03.013
Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex
Abstract
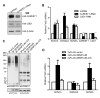
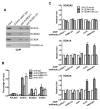
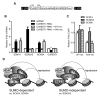
Posttranslational modification of transcription factors by the small ubiquitin-related modifier SUMO is associated with transcriptional repression, but the underlying mechanisms remain incompletely described. We have identified binding of the LSD1/CoREST1/HDAC corepressor complex to SUMO-2. Here we show that CoREST1 binds directly and noncovalently to SUMO-2, but not SUMO-1, and CoREST1 bridges binding of the histone demethylase LSD1 to SUMO-2. Depletion of SUMO-2/3 conjugates led to transcriptional derepression, reduced occupancy of CoREST1 and LSD1, and changes in histone methylation and acetylation at some, but not all, LSD1/CoREST1/HDAC target genes. We have identified a nonconsensus SUMO-interaction motif (SIM) in CoREST1 required for SUMO-2 binding, and we show that mutation of the CoREST1 SIM disrupted SUMO-2 binding and transcriptional repression of some neuronal-specific genes in nonneuronal cells. Our results reveal that direct interactions between CoREST1 and SUMO-2 mediate SUMO-dependent changes in chromatin structure and transcription that are important for cell-type-specific gene expression.
Figures




References
-
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. - PubMed
-
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. - PubMed
-
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

