FTIR analysis of GPCR activation using azido probes
- PMID: 19396177
- PMCID: PMC2875874
- DOI: 10.1038/nchembio.167
FTIR analysis of GPCR activation using azido probes
Abstract
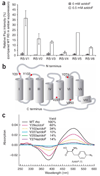
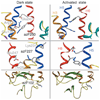
We demonstrate the site-directed incorporation of an IR-active amino acid, p-azido-L-phenylalanine (azidoF, 1), into the G protein-coupled receptor rhodopsin using amber codon suppression technology. The antisymmetric stretch vibration of the azido group absorbs at approximately 2,100 cm(-1) in a clear spectral window and is sensitive to its electrostatic environment. We used FTIR difference spectroscopy to monitor the azido probe and show that the electrostatic environments of specific interhelical networks change during receptor activation.
Figures

References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

