Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance
- PMID: 19398554
- PMCID: PMC2740416
- DOI: 10.1074/jbc.M808141200
Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance
Abstract
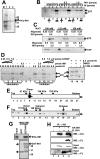
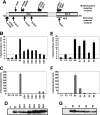
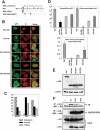
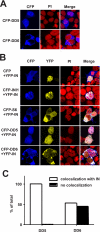
INI1/hSNF5/BAF47/SMARCB1 is an HIV-1 integrase (IN)-binding protein that modulates viral replication in multiple ways. A minimal IN-binding domain of INI1, S6 (amino acids 183-294), transdominantly inhibits late events, and down-modulation of INI1 stimulates early events of HIV-1 replication. INI1 both stimulates and inhibits in vitro integration depending on IN concentration. To gain further insight into its role in HIV-1 replication, we purified and biochemically characterized INI1. We found that INI1 forms multimeric structures. Deletion analysis indicated that the Rpt1 and Rpt2 motifs form the minimal multimerization domain. We isolated mutants of INI1 that are defective for multimerization using a reverse yeast two-hybrid system. Our results revealed that INI1 residues involved in multimerization overlap with IN-binding and nuclear export domains and are required for nuclear retention and co-localization with IN. Multimerization-defective mutants are also defective for mediating the transdominant effect of INI1-S6-(183-294). Furthermore, we found that INI1 is a minor groove DNA-binding protein. Although IN binding and multimerization are required for INI1-mediated inhibition, the acceptor DNA binding property of INI1 may be required for stimulation of in vitro strand transfer activities of IN. Binding of INI1 to IN results in the formation of presumably inactive high molecular weight IN-INI1 complexes, and the multimerization-defective mutant was unable to form these complexes. These results indicate that the multimerization and IN binding properties of INI1 are necessary for its ability to both inhibit integration and influence assembly and particle production, providing insights into the mechanism of INI1-mediated effects in HIV-1 replication.
Figures


References
-
- Sorin M., Kalpana G. V. (2006) Curr. HIV Res. 4, 117–130 - PubMed
-
- Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Science 319, 921–926 - PubMed
-
- Kalpana G. V., Marmon S., Wang W., Crabtree G. R., Goff S. P. (1994) Science 266, 2002–2006 - PubMed
-
- Van Maele B., Busschots K., Vandekerckhove L., Christ F., Debyser Z. (2006) Trends Biochem. Sci. 31, 98–105 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

