Identifying hubs in protein interaction networks
- PMID: 19399170
- PMCID: PMC2670494
- DOI: 10.1371/journal.pone.0005344
Identifying hubs in protein interaction networks
Abstract
Background: In spite of the scale-free degree distribution that characterizes most protein interaction networks (PINs), it is common to define an ad hoc degree scale that defines "hub" proteins having special topological and functional significance. This raises the concern that some conclusions on the functional significance of proteins based on network properties may not be robust.
Methodology: In this paper we present three objective methods to define hub proteins in PINs: one is a purely topological method and two others are based on gene expression and function. By applying these methods to four distinct PINs, we examine the extent of agreement among these methods and implications of these results on network construction.
Conclusions: We find that the methods agree well for networks that contain a balance between error-free and unbiased interactions, indicating that the hub concept is meaningful for such networks.
Conflict of interest statement
Figures






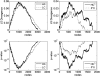
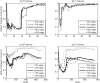
References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. - PubMed
-
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. - PubMed
-
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

