Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5' phosphate
- PMID: 19399178
- PMCID: PMC2670506
- DOI: 10.1371/journal.pone.0005347
Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5' phosphate
Abstract
Background: Holliday junction (HJ) resolution is a critical step during homologous recombination. In Escherichia coli this job is performed by a member of the RNase H/Integrase superfamily called RuvC, whereas in Schizosaccharomyces pombe it has been attributed to the XPF family member Mus81-Eme1. HJ resolution is achieved through the sequential cleavage of two strands of like polarity at or close to the junction crossover point. RuvC functions as a dimer, whereas Mus81-Eme1 is thought to function as a dimer of heterodimers. However, in both cases the multimer contains two catalytic sites, which act independently and sequentially during the resolution reaction. To ensure that both strands are cleaved before the nuclease dissociates from the junction, the rate of second strand cleavage is greatly enhanced compared to that of the first. The enhancement of second strand cleavage has been attributed to the increased flexibility of the nicked HJ, which would facilitate rapid engagement of the second active site and scissile bond. Here we have investigated whether other properties of the nicked HJ are important for enhancing second strand cleavage.
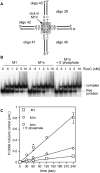
Principal findings: A comparison of the efficiency of cleavage of nicked HJs with and without a 5' phosphate at the nick site shows that a 5' phosphate is required for most of the enhancement of second strand cleavage by RuvC. In contrast Mus81-Eme1 cleaves nicked HJs with and without a 5' phosphate with equal efficiency, albeit there are differences in cleavage site selection.
Conclusions: Our data show that efficient HJ resolution by RuvC depends on the 5' phosphate revealed by incision of the first strand. This is a hitherto unappreciated factor in promoting accelerated second strand cleavage. However, a 5' phosphate is not a universal requirement since efficient cleavage by Mus81-Eme1 appears to depend solely on the increased junction flexibility that is developed by the first incision.
Conflict of interest statement
Figures


References
-
- Lilley DM, White MF. The junction-resolving enzymes. Nat Rev Mol Cell Biol. 2001;2:433–443. - PubMed
-
- Sharples GJ. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol Microbiol. 2001;39:823–834. - PubMed
-
- Declais AC, Lilley DM. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol. 2008;18:86–95. - PubMed
-
- Shah R, Bennett RJ, West SC. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994;79:853–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

