Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence
- PMID: 19399587
- PMCID: PMC2724633
- DOI: 10.1007/s00018-009-0029-z
Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence
Abstract
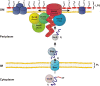
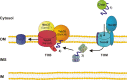
Membrane-embedded beta-barrel proteins span the membrane via multiple amphipathic beta-strands arranged in a cylindrical shape. These proteins are found in the outer membranes of Gram-negative bacteria, mitochondria and chloroplasts. This situation is thought to reflect the evolutionary origin of mitochondria and chloroplasts from Gram-negative bacterial endosymbionts. beta-barrel proteins fulfil a variety of functions; among them are pore-forming proteins that allow the flux of metabolites across the membrane by passive diffusion, active transporters of siderophores, enzymes, structural proteins, and proteins that mediate protein translocation across or insertion into membranes. The biogenesis process of these proteins combines evolutionary conservation of the central elements with some noticeable differences in signals and machineries. This review summarizes our current knowledge of the functions and biogenesis of this special family of proteins.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

