Escherichia coli DnaA forms helical structures along the longitudinal cell axis distinct from MreB filaments
- PMID: 19400775
- PMCID: PMC2888790
- DOI: 10.1111/j.1365-2958.2009.06674.x
Escherichia coli DnaA forms helical structures along the longitudinal cell axis distinct from MreB filaments
Abstract
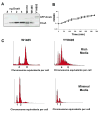
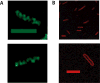
DnaA initiates chromosomal replication in Escherichia coli at a well-regulated time in the cell cycle. To determine how the spatial distribution of DnaA is related to the location of chromosomal replication and other cell cycle events, the localization of DnaA in living cells was visualized by confocal fluorescence microscopy. The gfp gene was randomly inserted into a dnaA-bearing plasmid via in vitro transposition to create a library that included internally GFP-tagged DnaA proteins. The library was screened for the ability to rescue dnaA(ts) mutants, and a candidate gfp-dnaA was used to replace the dnaA gene of wild-type cells. The resulting cells produce close to physiological levels of GFP-DnaA from the endogenous promoter as their only source of DnaA and somewhat under-initiate replication with moderate asynchrony. Visualization of GFP-tagged DnaA in living cells revealed that DnaA adopts a helical pattern that spirals along the long axis of the cell, a pattern also seen in wild-type cells by immunofluorescence with affinity purified anti-DnaA antibody. Although the DnaA helices closely resemble the helices of the actin analogue MreB, co-visualization of GFP-tagged DnaA and RFP-tagged MreB demonstrates that DnaA and MreB adopt discrete helical structures along the length of the longitudinal cell axis.
Figures


References
-
- Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem. 2007;282:17816–17827. - PubMed
-
- Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J Mol Biol. 2006;356:850–863. - PubMed
-
- Aranovich A, Gdalevsky GY, Cohen-Luria R, Fishov I, Parola AH. Membrane-catalyzed nucleotide exchange on DnaA. Effect of surface molecular crowding. J Biol Chem. 2006;281:12526–12534. - PubMed
-
- Aranovich A, Parola AH, Fishov I. The reactivation of DnaAL366K requires less acidic phospholipids supporting their role in the initiation of chromosome replication in Escherichia coli. FEBS letters. 2007;581:4439–4442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

