Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis
- PMID: 19400954
- PMCID: PMC2685784
- DOI: 10.1186/1743-422X-6-44
Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis
Abstract
Background: Proteins associated with the late endosome (LE) appear to play a central role in the envelopment of a number of taxonomically diverse viruses. How viral proteins interact with LE-associated proteins to facilitate envelopment is not well understood. LE-derived transport vesicles form through the interaction of Rab9 GTPase with cargo proteins, and TIP47, a Rab9-specific effector protein. Vaccinia virus (VV) induces a wrapping complex derived from intracellular host membranes to envelope intracellular mature virus particles producing egress-competent forms of virus.
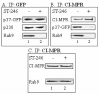
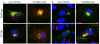
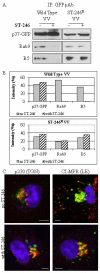
Results: We show that VV p37 protein associates with TIP47-, Rab9-, and CI-MPR-containing membranes. Mutation of a di-aromatic motif in p37 blocks association with TIP47 and inhibits plaque formation. ST-246, a specific inhibitor of p37 function, inhibits these interactions and also blocks wrapped virus particle formation. Vaccinia virus expressing p37 variants with reduced ST-246 susceptibility associates with Rab9 and co-localizes with CI-MPR in the presence and absence of compound.
Conclusion: These results suggest that p37 localizes to the LE and interacts with proteins associated with LE-derived transport vesicle biogenesis to facilitate assembly of extracellular forms of virus.
Figures




References
-
- Smith GL, Vanderplasschen A, Law M. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol. 2002;83:2915–2931. - PubMed
-
- Moss B. Poxviridae and their replication. In: Knipe B, Howley P, editor. Fields Virology. New York: Raven Press; 2001. pp. 2849–2884.
-
- Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

