Tamoxifen-elicited uterotrophy: cross-species and cross-ligand analysis of the gene expression program
- PMID: 19400957
- PMCID: PMC2683873
- DOI: 10.1186/1755-8794-2-19
Tamoxifen-elicited uterotrophy: cross-species and cross-ligand analysis of the gene expression program
Abstract
Background: Tamoxifen (TAM) is a well characterized breast cancer drug and selective estrogen receptor modulator (SERM) which also has been associated with a small increase in risk for uterine cancers. TAM's partial agonist activation of estrogen receptor has been characterized for specific gene promoters but not at the genomic level in vivo.Furthermore, reducing uncertainties associated with cross-species extrapolations of pharmaco- and toxicogenomic data remains a formidable challenge.
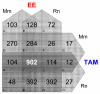
Results: A comparative ligand and species analysis approach was conducted to systematically assess the physiological, morphological and uterine gene expression alterations elicited across time by TAM and ethynylestradiol (EE) in immature ovariectomized Sprague-Dawley rats and C57BL/6 mice. Differential gene expression was evaluated using custom cDNA microarrays, and the data was compared to identify conserved and divergent responses. 902 genes were differentially regulated in all four studies, 398 of which exhibit identical temporal expression patterns.
Conclusion: Comparative analysis of EE and TAM differentially expressed gene lists suggest TAM regulates no unique uterine genes that are conserved in the rat and mouse. This demonstrates that the partial agonist activities of TAM extend to molecular targets in regulating only a subset of EE-responsive genes. Ligand-conserved, species-divergent expression of carbonic anhydrase 2 was observed in the microarray data and confirmed by real time PCR. The identification of comparable temporal phenotypic responses linked to related gene expression profiles demonstrates that systematic comparative genomic assessments can elucidate important conserved and divergent mechanisms in rodent estrogen signalling during uterine proliferation.
Figures




Similar articles
-
o-p'-DDT-mediated uterotrophy and gene expression in immature C57BL/6 mice and Sprague-Dawley rats.Toxicol Appl Pharmacol. 2013 Dec 15;273(3):532-41. doi: 10.1016/j.taap.2013.09.024. Epub 2013 Oct 3. Toxicol Appl Pharmacol. 2013. PMID: 24096037
-
Comparative temporal and dose-dependent morphological and transcriptional uterine effects elicited by tamoxifen and ethynylestradiol in immature, ovariectomized mice.BMC Genomics. 2007 Jun 7;8:151. doi: 10.1186/1471-2164-8-151. BMC Genomics. 2007. PMID: 17555576 Free PMC article.
-
Effects of tamoxifen and ethynylestradiol cotreatment on uterine gene expression in immature, ovariectomized mice.J Mol Endocrinol. 2010 Sep;45(3):161-73. doi: 10.1677/JME-09-0158. Epub 2010 Jul 13. J Mol Endocrinol. 2010. PMID: 20628019
-
Improving the efficacy of hormone therapy in breast cancer: The role of cholesterol metabolism in SERM-mediated autophagy, cell differentiation and death.Biochem Pharmacol. 2017 Nov 15;144:18-28. doi: 10.1016/j.bcp.2017.06.120. Epub 2017 Jun 19. Biochem Pharmacol. 2017. PMID: 28642035 Review.
-
Tamoxifen Resistance: Emerging Molecular Targets.Int J Mol Sci. 2016 Aug 19;17(8):1357. doi: 10.3390/ijms17081357. Int J Mol Sci. 2016. PMID: 27548161 Free PMC article. Review.
Cited by
-
Comparisons of differential gene expression elicited by TCDD, PCB126, βNF, or ICZ in mouse hepatoma Hepa1c1c7 cells and C57BL/6 mouse liver.Toxicol Lett. 2013 Oct 23;223(1):52-9. doi: 10.1016/j.toxlet.2013.08.013. Epub 2013 Aug 29. Toxicol Lett. 2013. PMID: 23994337 Free PMC article.
-
Endocrine Disrupting Chemicals and Endometrial Cancer: An Overview of Recent Laboratory Evidence and Epidemiological Studies.Int J Environ Res Public Health. 2017 Mar 22;14(3):334. doi: 10.3390/ijerph14030334. Int J Environ Res Public Health. 2017. PMID: 28327540 Free PMC article. Review.
-
Comparative uterotrophic effects of endoxifen and tamoxifen in ovariectomized Sprague-Dawley rats.Toxicol Pathol. 2014 Dec;42(8):1188-96. doi: 10.1177/0192623314525688. Epub 2014 Mar 26. Toxicol Pathol. 2014. PMID: 24670817 Free PMC article.
-
Skeletal and Uterotrophic Effects of Endoxifen in Female Rats.Endocrinology. 2017 Oct 1;158(10):3354-3368. doi: 10.1210/en.2016-1871. Endocrinology. 2017. PMID: 28977607 Free PMC article.
-
Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells.BMC Genomics. 2011 Apr 15;12:193. doi: 10.1186/1471-2164-12-193. BMC Genomics. 2011. PMID: 21496263 Free PMC article.
References
-
- Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, Wehling M. Nongenomic steroid action: controversies, questions, and answers. Physiol Rev. 2003;83:965–1016. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

