Highly efficient incorporation of the fluorescent nucleotide analogs tC and tCO by Klenow fragment
- PMID: 19401439
- PMCID: PMC2709563
- DOI: 10.1093/nar/gkp266
Highly efficient incorporation of the fluorescent nucleotide analogs tC and tCO by Klenow fragment
Abstract
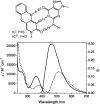
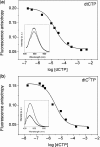
Studies of the mechanisms by which DNA polymerases select the correct nucleotide frequently employ fluorescently labeled DNA to monitor conformational rearrangements of the polymerase-DNA complex in response to incoming nucleotides. For this purpose, fluorescent base analogs play an increasingly important role because they interfere less with the DNA-protein interaction than do tethered fluorophores. Here we report the incorporation of the 5'-triphosphates of two exceptionally bright cytosine analogs, 1,3-diaza-2-oxo-phenothiazine (tC) and its oxo-homolog, 1,3-diaza-2-oxo-phenoxazine (tC(O)), into DNA by the Klenow fragment. Both nucleotide analogs are polymerized with slightly higher efficiency opposite guanine than cytosine triphosphate and are shown to bind with nanomolar affinity to the DNA polymerase active site, according to fluorescence anisotropy measurements. Using this method, we perform competitive binding experiments and show that they can be used to determine the dissociation constant of any given natural or unnatural nucleotide. The results demonstrate that the active site of the Klenow fragment is flexible enough to tolerate base pairs that are size-expanded in the major groove. In addition, the possibility to enzymatically polymerize a fluorescent nucleotide with high efficiency complements the tool box of biophysical probes available to study DNA replication.
Figures



Similar articles
-
High density labeling of polymerase chain reaction products with the fluorescent base analogue tCo.Anal Chem. 2009 Nov 1;81(21):9079-85. doi: 10.1021/ac9017555. Anal Chem. 2009. PMID: 19810708 Free PMC article.
-
Ambivalent incorporation of the fluorescent cytosine analogues tC and tCo by human DNA polymerase alpha and Klenow fragment.Biochemistry. 2009 Aug 11;48(31):7547-55. doi: 10.1021/bi9006995. Biochemistry. 2009. PMID: 19580325 Free PMC article.
-
Effect of transition metal ions on the fluorescence and Taq-catalyzed polymerase chain reaction of tricyclic cytidine analogs.Anal Biochem. 2011 Sep 1;416(1):53-60. doi: 10.1016/j.ab.2011.04.033. Epub 2011 Apr 27. Anal Biochem. 2011. PMID: 21600183 Free PMC article.
-
Influence of 5'-nearest neighbors on the insertion kinetics of the fluorescent nucleotide analog 2-aminopurine by Klenow fragment.Biochemistry. 1993 Oct 19;32(41):11247-58. doi: 10.1021/bi00092a039. Biochemistry. 1993. PMID: 8218190
-
Conformational dynamics of DNA polymerase probed with a novel fluorescent DNA base analogue.Biochemistry. 2007 Oct 30;46(43):12289-97. doi: 10.1021/bi700755m. Epub 2007 Oct 4. Biochemistry. 2007. PMID: 17915941
Cited by
-
Polymerase/DNA interactions and enzymatic activity: multi-parameter analysis with electro-switchable biosurfaces.Sci Rep. 2015 Jul 15;5:12066. doi: 10.1038/srep12066. Sci Rep. 2015. PMID: 26174478 Free PMC article.
-
High density labeling of polymerase chain reaction products with the fluorescent base analogue tCo.Anal Chem. 2009 Nov 1;81(21):9079-85. doi: 10.1021/ac9017555. Anal Chem. 2009. PMID: 19810708 Free PMC article.
-
Incorporation of the fluorescent ribonucleotide analogue tCTP by T7 RNA polymerase.Anal Chem. 2010 Feb 1;82(3):1082-9. doi: 10.1021/ac902456n. Anal Chem. 2010. PMID: 20067253 Free PMC article.
-
Templated 3' terminal fluorescent labeling of RNA using Klenow DNA polymerase.MethodsX. 2024 Aug 28;13:102925. doi: 10.1016/j.mex.2024.102925. eCollection 2024 Dec. MethodsX. 2024. PMID: 39290472 Free PMC article.
-
Structural and Molecular Kinetic Features of Activities of DNA Polymerases.Int J Mol Sci. 2022 Jun 7;23(12):6373. doi: 10.3390/ijms23126373. Int J Mol Sci. 2022. PMID: 35742812 Free PMC article. Review.
References
-
- Asseline U. Development and applications of fluorescent oligonucleotides. Curr. Org. Chem. 2006;10:491–518.
-
- Cekan P, Sigurdsson ST. Single base interrogation by a fluorescent nucleotide: each of the four DNA bases identified by fluorescence spectroscopy. Chem. Commun. 2008;29:3393–3395. - PubMed
-
- Tanaka K, Okamoto A. Design of a pyrene-containing fluorescence probe for labeling of RNA poly(A) tracts. Bioorg. Med. Chem. 2008;16:400–404. - PubMed

