Comparative proteomics analyses reveal the virB of B. melitensis affects expression of intracellular survival related proteins
- PMID: 19401764
- PMCID: PMC2670520
- DOI: 10.1371/journal.pone.0005368
Comparative proteomics analyses reveal the virB of B. melitensis affects expression of intracellular survival related proteins
Abstract
Background: Brucella melitensis is a facultative, intracellular, pathogenic bacterium that replicates within macrophages. The type IV secretion system encoded by the virB operon (virB) is involved in Brucella intracellular survival. However, the underlying molecular mechanisms, especially the target proteins affected by the virB, remain largely unclear.
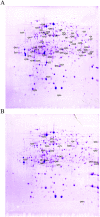
Methodology/principal findings: In order to define the proteins affected by virB, the proteomes of wild-type and the virB mutant were compared under in vitro conditions where virB was highly activated. The differentially expressed proteins were identified by MALDI-TOF-MS. Forty-four down-regulated and eighteen up-regulated proteins which exhibited a 2-fold or greater change were identified. These proteins included those involved in amino acid transport and metabolism, lipid metabolism, energy production, cell membrane biogenesis, translation, post-translational modifications and protein turnover, as well as unknown proteins. Interestingly, several important virulence related proteins involved in intracellular survival, including VjbR, DnaK, HtrA, Omp25, and GntR, were down-regulated in the virB mutant. Transcription analysis of virB and vjbR at different growth phase showed that virB positively affect transcription of vjbR in a growth phase dependent manner. Quantitative RT-PCR showed that transcription of these genes was also affected by virB during macrophage cell infection, consistent with the observed decreased survival of the virB mutant in macrophage.
Conclusions/significance: These data indicated that the virB operon may control the intracellular survival of Brucella by affecting the expression of relevant proteins.
Conflict of interest statement
Figures





References
-
- Boschiroli ML, Foulongne V, O'Callaghan D. Brucellosis: a worldwide zoonosis. Curr Opin Microbiol. 2001;4:58–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

