Selective enrichment of azide-containing peptides from complex mixtures
- PMID: 19402736
- PMCID: PMC2761887
- DOI: 10.1021/pr900257z
Selective enrichment of azide-containing peptides from complex mixtures
Abstract
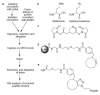
A general method is described to sequester peptides containing azides from complex peptide mixtures, aimed at facilitating mass spectrometric analysis to study different aspects of proteome dynamics. The enrichment method is based on covalent capture of azide-containing peptides by the azide-reactive cyclooctyne (ARCO) resin and is demonstrated for two different applications. Enrichment of peptides derived from cytochrome c treated with the azide-containing cross-linker bis(succinimidyl)-3-azidomethyl glutarate (BAMG) shows several cross-link containing peptides. Sequestration of peptides derived from an Escherichia coli proteome, pulse labeled with the bio-orthogonal amino acid azidohomoalanine as substitute for methionine, allows identification of numerous newly synthesized proteins. Furthermore, the method is found to be very specific, as after enrichment over 87% of all peptides contain (modified) azidohomoalanine.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

