Accelerated adaptive evolution on a newly formed X chromosome
- PMID: 19402745
- PMCID: PMC2672600
- DOI: 10.1371/journal.pbio.1000082
Accelerated adaptive evolution on a newly formed X chromosome
Abstract
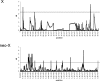
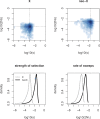
Sex chromosomes originated from ordinary autosomes, and their evolution is characterized by continuous gene loss from the ancestral Y chromosome. Here, we document a new feature of sex chromosome evolution: bursts of adaptive fixations on a newly formed X chromosome. Taking advantage of the recently formed neo-X chromosome of Drosophila miranda, we compare patterns of DNA sequence variation at genes located on the neo-X to genes on the ancestral X chromosome. This contrast allows us to draw inferences of selection on a newly formed X chromosome relative to background levels of adaptation in the genome while controlling for demographic effects. Chromosome-wide synonymous diversity on the neo-X is reduced 2-fold relative to the ancestral X, as expected under recent and recurrent directional selection. Several statistical tests employing various features of the data consistently identify 10%-15% of neo-X genes as targets of recent adaptive evolution but only 1%-3% of genes on the ancestral X. In addition, both the rate of adaptation and the fitness effects of adaptive substitutions are estimated to be roughly an order of magnitude higher for neo-X genes relative to genes on the ancestral X. Thus, newly formed X chromosomes are not passive players in the evolutionary process of sex chromosome differentiation, but respond adaptively to both their sex-biased transmission and to Y chromosome degeneration, possibly through demasculinization of their gene content and the evolution of dosage compensation.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

References
-
- Bull JJ. Evolution of sex determining mechanisms. Menlo Park (California): Benjamin Cummings; 1983. 316
-
- Skaletsky H, Kuroda-Kawaguchi T, Minx P, Cordum H, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. - PubMed
-
- Bachtrog D. Adaptation shapes patterns of genome evolution in sexual and asexual genomes in Drosophila. Nat Genet. 2003;34:215–219. - PubMed
-
- Guttman D, Charlesworth D. An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature. 1998;393:263–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

