Towards high-throughput molecular detection of Plasmodium: new approaches and molecular markers
- PMID: 19402894
- PMCID: PMC2686730
- DOI: 10.1186/1475-2875-8-86
Towards high-throughput molecular detection of Plasmodium: new approaches and molecular markers
Abstract
Background: Several strategies are currently deployed in many countries in the tropics to strengthen malaria control toward malaria elimination. To measure the impact of any intervention, there is a need to detect malaria properly. Mostly, decisions still rely on microscopy diagnosis. But sensitive diagnosis tools enabling to deal with a large number of samples are needed. The molecular detection approach offers a much higher sensitivity, and the flexibility to be automated and upgraded.
Methods: Two new molecular methods were developed: dot18S, a Plasmodium-specific nested PCR based on the 18S rRNA gene followed by dot-blot detection of species by using species-specific probes and CYTB, a Plasmodium-specific nested PCR based on cytochrome b gene followed by species detection using SNP analysis. The results were compared to those obtained with microscopic examination and the "standard" 18S rRNA gene based nested PCR using species specific primers. 337 samples were diagnosed.
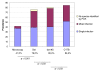
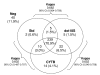
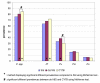
Results: Compared to the microscopy the three molecular methods were more sensitive, greatly increasing the estimated prevalence of Plasmodium infection, including P. malariae and P. ovale. A high rate of mixed infections was uncovered with about one third of the villagers infected with more than one malaria parasite species. Dot18S and CYTB sensitivity outranged the "standard" nested PCR method, CYTB being the most sensitive. As a consequence, compared to the "standard" nested PCR method for the detection of Plasmodium spp., the sensitivity of dot18S and CYTB was respectively 95.3% and 97.3%. Consistent detection of Plasmodium spp. by the three molecular methods was obtained for 83% of tested isolates. Contradictory results were mostly related to detection of Plasmodium malariae and Plasmodium ovale in mixed infections, due to an "all-or-none" detection effect at low-level parasitaemia.
Conclusion: A large reservoir of asymptomatic infections was uncovered using the molecular methods. Dot18S and CYTB, the new methods reported herein are highly sensitive, allow parasite DNA extraction as well as genus- and species-specific diagnosis of several hundreds of samples, and are amenable to high-throughput scaling up for larger sample sizes. Such methods provide novel information on malaria prevalence and epidemiology and are suited for active malaria detection. The usefulness of such sensitive malaria diagnosis tools, especially in low endemic areas where eradication plans are now on-going, is discussed in this paper.
Figures

References
-
- Barker RH, Jr, Suebsaeng L, Rooney W, Alecrim GC, Dourado HV, Wirth DF. Specific DNA probe for the diagnosis of Plasmodium falciparum malaria. Science. 1986;231:1434–1436. - PubMed
-
- Franzen L, Westin G, Shabo R, Aslund L, Perlmann H, Persson T, Wigzell H, Pettersson U. Analysis of clinical specimens by hybridisation with probe containing repetitive DNA from Plasmodium falciparum. A novel approach to malaria diagnosis. Lancet. 1984;1:525–528. - PubMed
-
- Pollack Y, Metzger S, Shemer R, Landau D, Spira DT, Golenser J. Detection of Plasmodium falciparum in blood using DNA hybridization. Am J Trop Med Hyg. 1985;34:663–667. - PubMed
-
- de Monbrison F, Angei C, Staal A, Kaiser K, Picot S. Simultaneous identification of the four human Plasmodium species and quantification of Plasmodium DNA load in human blood by real-time polymerase chain reaction. Trans R Soc Trop Med Hyg. 2003;97:387–390. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

