New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses
- PMID: 19402957
- PMCID: PMC2687021
- DOI: 10.3201/eid1505.081286
New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses
Abstract
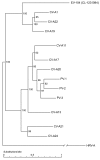
Rhinoviruses and enteroviruses are leading causes of respiratory infections. To evaluate genotypic diversity and identify forces shaping picornavirus evolution, we screened persons with respiratory illnesses by using rhinovirus-specific or generic real-time PCR assays. We then sequenced the 5 untranslated region, capsid protein VP1, and protease precursor 3CD regions of virus-positive samples. Subsequent phylogenetic analysis identified the large genotypic diversity of rhinoviruses circulating in humans. We identified and completed the genome sequence of a new enterovirus genotype associated with respiratory symptoms and acute otitis media, confirming the close relationship between rhinoviruses and enteroviruses and the need to detect both viruses in respiratory specimens. Finally, we identified recombinants among circulating rhinoviruses and mapped their recombination sites, thereby demonstrating that rhinoviruses can recombine in their natural host. This study clarifies the diversity and explains the reasons for evolution of these viruses.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
