Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly
- PMID: 19403693
- PMCID: PMC2695803
- DOI: 10.1091/mbc.e08-12-1251
Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly
Abstract
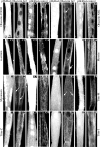
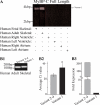
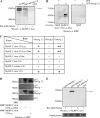
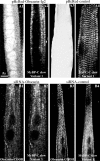
Obscurin is a multidomain protein composed of adhesion and signaling domains that plays key roles in the organization of contractile and membrane structures in striated muscles. Overexpression of the second immunoglobulin domain of obscurin (Ig2) in developing myotubes inhibits the assembly of A- and M-bands, but not Z-disks or I-bands. This effect is mediated by the direct interaction of the Ig2 domain of obscurin with a novel isoform of myosin binding protein-C slow (MyBP-C slow), corresponding to variant-1. Variant-1 contains all the structural motifs present in the known forms of MyBP-C slow, but it has a unique COOH terminus. Quantitative reverse transcription-polymerase chain reaction indicated that MyBP-C slow variant-1 is expressed in skeletal muscles both during development and at maturity. Immunolabeling of skeletal myofibers with antibodies to the unique COOH terminus of variant-1 demonstrated that, unlike other forms of MyBP-C slow that reside in the C-zones of A-bands, variant-1 preferentially concentrates around M-bands, where it codistributes with obscurin. Overexpression of the Ig2 domain of obscurin or reduction of expression of obscurin inhibited the integration of variant-1 into forming M-bands in skeletal myotubes. Collectively, our experiments identify a new ligand of obscurin at the M-band, MyBP-C slow variant-1 and suggest that their interaction contributes to the assembly of M- and A-bands.
Figures





References
-
- Alyonycheva T. N., Mikawa T., Reinach F. C., Fischman D. A. Isoform-specific interaction of the myosin-binding proteins (MyBPs) with skeletal and cardiac myosin is a property of the C-terminal immunoglobulin domain. J. Biol. Chem. 1997;272:20866–20872. - PubMed
-
- Arimura T., Matsumoto Y., Okazaki O., Hayashi T., Takahashi M., Inagaki N., Hinohara K., Ashizawa N., Yano K., Kimura A. Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2007;362:281–287. - PubMed
-
- Armani A., Galli S., Giacomello E., Bagnato P., Barone V., Rossi D., Sorrentino V. Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp. Cell Res. 2006;312:3546–3558. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

