SRC family kinases are required for limb trajectory selection by spinal motor axons
- PMID: 19403835
- PMCID: PMC6665840
- DOI: 10.1523/JNEUROSCI.0265-09.2009
SRC family kinases are required for limb trajectory selection by spinal motor axons
Abstract
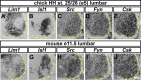
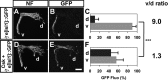
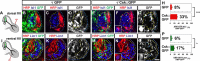
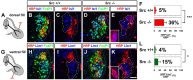
Signal relay by guidance receptors at the axonal growth cone is a process essential for the assembly of a functional nervous system. We investigated the in vivo function of Src family kinases (SFKs) as growth cone guidance signaling intermediates in the context of spinal lateral motor column (LMC) motor axon projection toward the ventral or dorsal limb mesenchyme. Using in situ mRNA detection we determined that Src and Fyn are expressed in LMC motor neurons of chick and mouse embryos at the time of limb trajectory selection. Inhibition of SFK activity by C-terminal Src kinase (Csk) overexpression in chick LMC axons using in ovo electroporation resulted in LMC axons selecting the inappropriate dorsoventral trajectory within the limb mesenchyme, with medial LMC axon projecting into the dorsal and ventral limb nerve with apparently random incidence. We also detected LMC axon trajectory choice errors in Src mutant mice demonstrating a nonredundant role for Src in motor axon guidance in agreement with gain and loss of Src function in chick LMC neurons which led to the redirection of LMC axons. Finally, Csk-mediated SFK inhibition attenuated the retargeting of LMC axons caused by EphA or EphB over-expression, implying the participation of SFKs in Eph-mediated LMC motor axon guidance. In summary, our findings demonstrate that SFKs are essential for motor axon guidance and suggest that they play an important role in relaying ephrin:Eph signals that mediate the selection of motor axon trajectory in the limb.
Figures


Similar articles
-
Ephexin1 Is Required for Eph-Mediated Limb Trajectory of Spinal Motor Axons.J Neurosci. 2018 Feb 21;38(8):2043-2056. doi: 10.1523/JNEUROSCI.2257-17.2018. Epub 2018 Jan 23. J Neurosci. 2018. PMID: 29363583 Free PMC article.
-
Paxillin Is Required for Proper Spinal Motor Axon Growth into the Limb.J Neurosci. 2021 Apr 28;41(17):3808-3821. doi: 10.1523/JNEUROSCI.2863-20.2021. Epub 2021 Mar 16. J Neurosci. 2021. PMID: 33727334 Free PMC article.
-
Nck2 is essential for limb trajectory selection by spinal motor axons.Dev Dyn. 2018 Sep;247(9):1043-1056. doi: 10.1002/dvdy.24656. Epub 2018 Sep 6. Dev Dyn. 2018. PMID: 30016580
-
Lateral motor column axons execute a ternary trajectory choice between limb and body tissues.Neural Dev. 2007 Jul 2;2:13. doi: 10.1186/1749-8104-2-13. Neural Dev. 2007. PMID: 17605791 Free PMC article.
-
Spontaneous rhythmic activity in early chick spinal cord influences distinct motor axon pathfinding decisions.Brain Res Rev. 2008 Jan;57(1):77-85. doi: 10.1016/j.brainresrev.2007.06.021. Epub 2007 Aug 1. Brain Res Rev. 2008. PMID: 17920131 Free PMC article. Review.
Cited by
-
The Geometry of Limb Motor Innervation is Controlled by the Dorsal-Ventral Compartment Boundary in the Chick Limbless Mutant.Neuroscience. 2020 Dec 1;450:29-47. doi: 10.1016/j.neuroscience.2020.09.054. Epub 2020 Oct 8. Neuroscience. 2020. PMID: 33038447 Free PMC article.
-
Emergence of motor circuit activity.PLoS One. 2014 Apr 10;9(4):e93836. doi: 10.1371/journal.pone.0093836. eCollection 2014. PLoS One. 2014. PMID: 24722186 Free PMC article.
-
Plasticity versus specificity in RTK signalling modalities for distinct biological outcomes in motor neurons.BMC Biol. 2014 Aug 14;12:56. doi: 10.1186/s12915-014-0056-6. BMC Biol. 2014. PMID: 25124859 Free PMC article.
-
Genetic analysis of DSCAM's role as a Netrin-1 receptor in vertebrates.J Neurosci. 2012 Jan 11;32(2):411-6. doi: 10.1523/JNEUROSCI.3563-11.2012. J Neurosci. 2012. PMID: 22238077 Free PMC article.
-
Motor axon pathfinding.Cold Spring Harb Perspect Biol. 2010 Mar;2(3):a001735. doi: 10.1101/cshperspect.a001735. Cold Spring Harb Perspect Biol. 2010. PMID: 20300210 Free PMC article. Review.
References
-
- Beg AA, Sommer JE, Martin JH, Scheiffele P. α2-Chimaerin is an essential EphA4 effector in the assembly of neuronal locomotor circuits. Neuron. 2007;55:768–778. - PubMed
-
- Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Vav family GEFs link activated Ephs to endocytosis and axon guidance. Neuron. 2005;46:205–217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
