Signal transducer and activator of transcription 1 (STAT1) is essential for chromium silencing of gene induction in human airway epithelial cells
- PMID: 19403854
- PMCID: PMC2696327
- DOI: 10.1093/toxsci/kfp084
Signal transducer and activator of transcription 1 (STAT1) is essential for chromium silencing of gene induction in human airway epithelial cells
Abstract
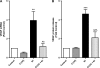
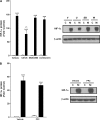
Hexavalent chromium (Cr(VI)) promotes lung injury and pulmonary diseases through poorly defined mechanisms that may involve the silencing of inducible protective genes. The current study investigated the hypothesis that Cr(VI) actively signals through a signal transducer and activator of transcription 1 (STAT1)-dependent pathway to silence nickel (Ni)-induced expression of vascular endothelial cell growth factor A (VEGFA), an important mediator of lung injury and repair. In human bronchial airway epithelial (BEAS-2B) cells, Ni-induced VEGFA transcription by stimulating an extracellular regulated kinase (ERK) signaling cascade that involved Src kinase-activated Sp1 transactivation, as well as increased hypoxia-inducible factor-1 alpha (HIF-1 alpha) stabilization and DNA binding. Ni-stimulated ERK, Src, and HIF-1 alpha activities, as well as Ni-induced VEGFA transcript levels were inhibited in Cr(VI)-exposed cells. We previously demonstrated that Cr(VI) stimulates STAT1 to suppress VEGFA expression. In BEAS-2B cells stably expressing STAT1 short hairpin RNA, Cr(VI) increased VEGFA transcript levels and Sp1 transactivation. Moreover, in the absence of STAT1, Cr(VI), and Ni coexposures positively interacted to further increase VEGFA transcripts. This study demonstrates that metal-stimulated signaling cascades interact to regulate transcription and induction of adaptive or repair responses in airway cells. In addition, the data implicate STAT1 as a rate limiting mediator of Cr(VI)-stimulated gene regulation and suggest that cells lacking STAT1, such as many tumor cell lines, have opposite responses to Cr(VI) relative to normal cells.
Figures





References
-
- Acarregui MJ, Penisten ST, Goss KL, Ramirez K, Snyder JM. Vascular endothelial growth factor gene expression in human fetal lung in vitro. Am. J. Respir. Cell Mol. Biol. 1999;20(1):14–23. - PubMed
-
- Andrew AS, Klei LR, Barchowsky A. Nickel requires hypoxia-inducible factor-1α, not redox signaling to induce plasminogen activator inhibitor-1. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;281:L607–L615. - PubMed
-
- Antonini JM, Stone S, Roberts JR, Chen B, Schwegler-Berry D, Afshari AA, Frazer DG. Effect of short-term stainless steel welding fume inhalation exposure on lung inflammation, injury, and defense responses in rats. Toxicol. Appl. Pharmacol. 2007;223(3):234–245. - PubMed
-
- Antonini JM, Taylor MD, Zimmer AT, Roberts JR. Pulmonary responses to welding fumes: Role of metal constituents. J. Toxicol. Environ. Health A. 2004;67(3):233–249. - PubMed
-
- Barchowsky A, Roussel RR, Krieser RJ, Mossman BT, Treadwell MD. Expression and activity of urokinase and its receptor in endothelial and pulmonary epithelial cells exposed to asbestos. Toxicol. Appl. Pharmacol. 1998;152(2):388–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

