Latent evolutionary potentials under the neutral mutational drift of an enzyme
- PMID: 19404461
- PMCID: PMC2645560
- DOI: 10.2976/1.2739115/10.2976/1
Latent evolutionary potentials under the neutral mutational drift of an enzyme
Abstract
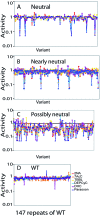
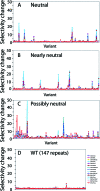
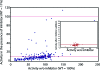
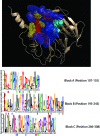
Biological systems exhibit mutational robustness, or neutrality, whereby the impact of mutations is minimized. Does neutrality hamper their ability to adapt in the face of changing environments? We monitored changes in genotype and phenotype that occur within a neutral mutational network of an enzyme, experimentally and computationally (see accompanying article). Using the enzyme PON1 as a model, we performed random mutagenesis and purifying selection to purge deleterious mutations. We characterized approximately 300 variants that are apparently neutral, or close to neutral, with respect to PON1's levels of expression and native lactonase activity. Their activities with promiscuous substrates and ligands indicated significant changes in adaptive potentials. Almost half of the variants exhibited changes in promiscuous activities, specificities, or inhibition, and several of these were found to be one or two mutations, closer to potentially new phenotypes: aryl esterase, thiolactonase, phosphotriesterase, or drug resistance. This empirical measure of phenotypic changes under neutrality supports the notion that sequence changes that are neutral, i.e., non-adaptive, in a current context can facilitate adaptation under changing circumstances, by both expanding the activity range of existing enzymes and thus providing an immediate advantage, and by reducing the number of mutations required for divergence of new functions.
Figures


References
-
- Aharoni, A, Gaidukov, L, Khersonsky, O, Mc, Q GS, Roodveldt, C, and Tawfik, D S (2005b). “The ‘evolvability’ of promiscuous protein functions.” Nat. Genet. NGENEC 37, 73–76. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
