From DNA nanotechnology to synthetic biology
- PMID: 19404476
- PMCID: PMC2645571
- DOI: 10.2976/1.2896331
From DNA nanotechnology to synthetic biology
Abstract
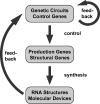
Attempts to construct artificial systems from biological molecules such as DNA and RNA by self-assembly are compatible with the recent development of synthetic biology. Genetic mechanisms can be used to produce or control artificial structures made from DNA and RNA, and these structures can in turn be used as artificial gene regulatory elements, in vitro as well as in vivo. Artificial biochemical circuits can be incorporated into cell-like reaction compartments, which opens up the possibility to operate them permanently out of equilibrium. In small systems, stochastic effects become noticeable and will have to be accounted for in the design of future systems.
Figures




References
LinkOut - more resources
Full Text Sources
