In silico miRNA prediction in metazoan genomes: balancing between sensitivity and specificity
- PMID: 19405940
- PMCID: PMC2688010
- DOI: 10.1186/1471-2164-10-204
In silico miRNA prediction in metazoan genomes: balancing between sensitivity and specificity
Abstract
Background: MicroRNAs (miRNAs), short approximately 21-nucleotide RNA molecules, play an important role in post-transcriptional regulation of gene expression. The number of known miRNA hairpins registered in the miRBase database is rapidly increasing, but recent reports suggest that many miRNAs with restricted temporal or tissue-specific expression remain undiscovered. Various strategies for in silico miRNA identification have been proposed to facilitate miRNA discovery. Notably support vector machine (SVM) methods have recently gained popularity. However, a drawback of these methods is that they do not provide insight into the biological properties of miRNA sequences.
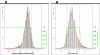
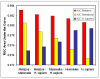
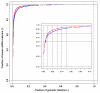
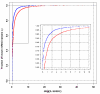
Results: We here propose a new strategy for miRNA hairpin prediction in which the likelihood that a genomic hairpin is a true miRNA hairpin is evaluated based on statistical distributions of observed biological variation of properties (descriptors) of known miRNA hairpins. These distributions are transformed into a single and continuous outcome classifier called the L score. Using a dataset of known miRNA hairpins from the miRBase database and an exhaustive set of genomic hairpins identified in the genome of Caenorhabditis elegans, a subset of 18 most informative descriptors was selected after detailed analysis of correlation among and discriminative power of individual descriptors. We show that the majority of previously identified miRNA hairpins have high L scores, that the method outperforms miRNA prediction by threshold filtering and that it is more transparent than SVM classifiers.
Conclusion: The L score is applicable as a prediction classifier with high sensitivity for novel miRNA hairpins. The L-score approach can be used to rank and select interesting miRNA hairpin candidates for downstream experimental analysis when coupled to a genome-wide set of in silico-identified hairpins or to facilitate the analysis of large sets of putative miRNA hairpin loci obtained in deep-sequencing efforts of small RNAs. Moreover, the in-depth analyses of miRNA hairpins descriptors preceding and determining the L score outcome could be used as an extension to miRBase entries to help increase the reliability and biological relevance of the miRNA registry.
Figures




Similar articles
-
MiRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans.BMC Bioinformatics. 2007 Sep 17;8:341. doi: 10.1186/1471-2105-8-341. BMC Bioinformatics. 2007. PMID: 17868480 Free PMC article.
-
Ensemble-based classification approach for micro-RNA mining applied on diverse metagenomic sequences.BMC Res Notes. 2014 May 6;7:286. doi: 10.1186/1756-0500-7-286. BMC Res Notes. 2014. PMID: 24884968 Free PMC article.
-
Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine.BMC Bioinformatics. 2005 Dec 29;6:310. doi: 10.1186/1471-2105-6-310. BMC Bioinformatics. 2005. PMID: 16381612 Free PMC article.
-
Functional genomic, computational and proteomic analysis of C. elegans microRNAs.Brief Funct Genomic Proteomic. 2008 May;7(3):228-35. doi: 10.1093/bfgp/eln024. Epub 2008 Jun 19. Brief Funct Genomic Proteomic. 2008. PMID: 18565984 Review.
-
The current landscape of microRNAs (miRNAs) in bacterial pneumonia: opportunities and challenges.Cell Mol Biol Lett. 2022 Aug 19;27(1):70. doi: 10.1186/s11658-022-00368-y. Cell Mol Biol Lett. 2022. PMID: 35986232 Free PMC article. Review.
Cited by
-
Drosha processing controls the specificity and efficiency of global microRNA expression.Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):700-7. doi: 10.1016/j.bbagrm.2011.05.015. Epub 2011 Jun 13. Biochim Biophys Acta. 2011. PMID: 21683814 Free PMC article.
-
Mirtrons: microRNA biogenesis via splicing.Biochimie. 2011 Nov;93(11):1897-904. doi: 10.1016/j.biochi.2011.06.017. Epub 2011 Jun 21. Biochimie. 2011. PMID: 21712066 Free PMC article. Review.
-
Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans.Genome Res. 2011 Feb;21(2):286-300. doi: 10.1101/gr.113050.110. Epub 2010 Dec 22. Genome Res. 2011. PMID: 21177960 Free PMC article.
-
Discovery of hundreds of mirtrons in mouse and human small RNA data.Genome Res. 2012 Sep;22(9):1634-45. doi: 10.1101/gr.133553.111. Genome Res. 2012. PMID: 22955976 Free PMC article.
-
Computational methods for ab initio detection of microRNAs.Front Genet. 2012 Oct 10;3:209. doi: 10.3389/fgene.2012.00209. eCollection 2012. Front Genet. 2012. PMID: 23087705 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

