Broadly targeted multiprobe QPCR for detection of coronaviruses: Coronavirus is common among mallard ducks (Anas platyrhynchos)
- PMID: 19406168
- PMCID: PMC7112901
- DOI: 10.1016/j.jviromet.2009.04.022
Broadly targeted multiprobe QPCR for detection of coronaviruses: Coronavirus is common among mallard ducks (Anas platyrhynchos)
Abstract
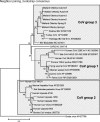
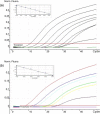
Coronaviruses (CoVs) can cause trivial or fatal disease in humans and in animals. Detection methods for a wide range of CoVs are needed, to understand viral evolution, host range, transmission and maintenance in reservoirs. A new concept, "Multiprobe QPCR", which uses a balanced mixture of competing discrete non- or moderately degenerated nuclease degradable (TaqMan) probes was employed. It provides a broadly targeted and rational single tube real-time reverse transcription PCR ("NQPCR") for the generic detection and discovery of CoV. Degenerate primers, previously published, and the new probes, were from a conserved stretch of open reading frame 1b, encoding the replicase. This multiprobe design reduced the degree of probe degeneration, which otherwise decreases the sensitivity, and allowed a preliminary classification of the amplified sequence directly from the QPCR trace. The split probe strategy allowed detection of down to 10 viral nucleic acid equivalents of CoV from all known CoV groups. Evaluation was with reference CoV strains, synthetic targets, human respiratory samples and avian fecal samples. Infectious-Bronchitis-Virus (IBV)-related variants were found in 7 of 35 sample pools, from 100 wild mallards (Anas platyrhynchos). Ducks may spread and harbour CoVs. NQPCR can detect a wide range of CoVs, as illustrated for humans and ducks.
Figures

References
-
- Adzhar A.B., Shaw K., Britton P., Cavanagh D. Avian infectious bronchitis virus: differences between 793/B and other strains. Vet. Rec. 1995;136:548. - PubMed
-
- Bohl E.H., Gupta R.K., McCloskey L.W., Saif L. Immunology of transmissible gastroenteritis. J. Am. Vet. Med. Assoc. 1972;160:543–549. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

