Genomic DNA microarray comparison of gene expression patterns in Paracoccidioides brasiliensis mycelia and yeasts in vitro
- PMID: 19406900
- PMCID: PMC2888123
- DOI: 10.1099/mic.0.027441-0
Genomic DNA microarray comparison of gene expression patterns in Paracoccidioides brasiliensis mycelia and yeasts in vitro
Abstract
Paracoccidioides brasiliensis is a thermally dimorphic fungus, and causes the most prevalent systemic mycosis in Latin America. Infection is initiated by inhalation of conidia or mycelial fragments by the host, followed by further differentiation into the yeast form. Information regarding gene expression by either form has rarely been addressed with respect to multiple time points of growth in culture. Here, we report on the construction of a genomic DNA microarray, covering approximately 25 % of the genome of the organism, and its utilization in identifying genes and gene expression patterns during growth in vitro. Cloned, amplified inserts from randomly sheared genomic DNA (gDNA) and known control genes were printed onto glass slides to generate a microarray of over 12,000 elements. To examine gene expression, mRNA was extracted and amplified from mycelial or yeast cultures grown in semi-defined medium for 5, 8 and 14 days. Principal components analysis and hierarchical clustering indicated that yeast gene expression profiles differed greatly from those of mycelia, especially at earlier time points, and that mycelial gene expression changed less than gene expression in yeasts over time. Genes upregulated in yeasts were found to encode proteins shown to be involved in methionine/cysteine metabolism, respiratory and metabolic processes (of sugars, amino acids, proteins and lipids), transporters (small peptides, sugars, ions and toxins), regulatory proteins and transcription factors. Mycelial genes involved in processes such as cell division, protein catabolism, nucleotide biosynthesis and toxin and sugar transport showed differential expression. Sequenced clones were compared with Histoplasma capsulatum and Coccidioides posadasii genome sequences to assess potentially common pathways across species, such as sulfur and lipid metabolism, amino acid transporters, transcription factors and genes possibly related to virulence. We also analysed gene expression with time in culture and found that while transposable elements and components of respiratory pathways tended to increase in expression with time, genes encoding ribosomal structural proteins and protein catabolism tended to sharply decrease in expression over time, particularly in yeast. These findings expand our knowledge of the different morphological forms of P. brasiliensis during growth in culture.
Figures
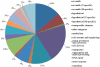



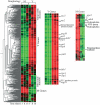
References
-
- Albuquerque, P., Kyaw, C. M., Saldanha, R. R., Brigido, M. M., Felipe, M. S. & Silva-Pereira, I. (2004). Pbhyd1 and Pbhyd2: two mycelium-specific hydrophobin genes from the dimorphic fungus Paracoccidioides brasiliensis. Fungal Genet Biol 41, 510–520. - PubMed
-
- Almeida, A. J., Matute, D. R., Carmona, J. A., Martins, M., Torres, I., McEwen, J. G., Restrepo, A., Leão, C., Ludovico, P. & Rodrigues, F. (2007). Genome size and ploidy of Paracoccidioides brasiliensis reveals a haploid DNA content: flow cytometry and GP43 sequence analysis. Fungal Genet Biol 44, 25–31. - PubMed
-
- Arraes, F. B., Benoliel, B., Burtet, R. T., Costa, P. L., Galdino, A. S., Lima, L. H., Marinho-Silva, C., Oliveira-Pereira, L., Pfrimer, P. & other authors (2005). General metabolism of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis. Genet Mol Res 4, 290–308. - PubMed
-
- Bagagli, E., Sano, A., Coelho, K. I., Alquati, S., Miyaji, M., de Camargo, Z. P., Gomes, G. M., Franco, M. & Montenegro, M. R. (1998). Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus noveminctus) captured in an endemic area of paracoccidioidomycosis. Am J Trop Med Hyg 58, 505–512. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

