Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach
- PMID: 19409270
- PMCID: PMC3632349
- DOI: 10.1016/j.neuron.2009.02.027
Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach
Abstract
The precise subunit composition of synaptic ionotropic receptors in the brain is poorly understood. This information is of particular importance with regard to AMPA-type glutamate receptors, the multimeric complexes assembled from GluA1-A4 subunits, as the trafficking of these receptors into and out of synapses is proposed to depend upon the subunit composition of the receptor. We report a molecular quantification of synaptic AMPA receptors (AMPARs) by employing a single-cell genetic approach coupled with electrophysiology in hippocampal CA1 pyramidal neurons. In contrast to prevailing views, we find that GluA1A2 heteromers are the dominant AMPARs at CA1 cell synapses (approximately 80%). In cells lacking GluA1, -A2, and -A3, synapses are devoid of AMPARs, yet synaptic NMDA receptors (NMDARs) and dendritic morphology remain unchanged. These data demonstrate a functional dissociation of AMPARs from trafficking of NMDARs and neuronal morphogenesis. This study provides a functional quantification of the subunit composition of AMPARs in the CNS and suggests novel roles for AMPAR subunits in receptor trafficking.
Figures

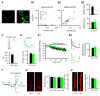
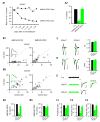
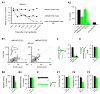
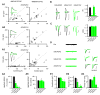
Comment in
-
AMPA receptor subunits get their share of the pie.Neuron. 2009 Apr 30;62(2):165-8. doi: 10.1016/j.neuron.2009.04.016. Neuron. 2009. PMID: 19409261
References
-
- Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. - PubMed
-
- Ayalon G, Segev E, Elgavish S, Stern-Bach Y. Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J Biol Chem. 2005;280:15053–15060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

