Phylogeny and virulence of naturally occurring type III secretion system-deficient Pectobacterium strains
- PMID: 19411432
- PMCID: PMC2704834
- DOI: 10.1128/AEM.01336-08
Phylogeny and virulence of naturally occurring type III secretion system-deficient Pectobacterium strains
Abstract
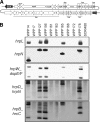
Pectobacterium species are enterobacterial plant-pathogenic bacteria that cause soft rot disease in diverse plant species. Previous epidemiological studies of Pectobacterium species have suffered from an inability to identify most isolates to the species or subspecies level. We used three previously described DNA-based methods, 16S-23S intergenic transcribed spacer PCR-restriction fragment length polymorphism analysis, multilocus sequence analysis (MLSA), and pulsed-field gel electrophoresis, to examine isolates from diseased stems and tubers and found that MLSA provided the most reliable classification of isolates. We found that strains belonging to at least two Pectobacterium clades were present in each field examined, although representatives of only three of five Pectobacterium clades were isolated. Hypersensitive response and DNA hybridization assays revealed that strains of both Pectobacterium carotovorum and Pectobacterium wasabiae lack a type III secretion system (T3SS). Two of the T3SS-deficient strains assayed lack genes adjacent to the T3SS gene cluster, suggesting that multiple deletions occurred in Pectobacterium strains in this locus, and all strains appear to have only six rRNA operons instead of the seven operons typically found in Pectobacterium strains. The virulence of most of the T3SS-deficient strains was similar to that of T3SS-encoding strains in stems and tubers.
Figures






References
-
- Bauer, D. W., A. J. Bogdanove, S. V. Beer, and A. Collmer. 1994. Erwinia chrysanthemi hrp genes, and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol. Plant-Microbe Interact. 7:573-581. - PubMed
-
- Bell, K. S., M. Sebaihia, L. Pritchard, M. T. G. Holden, L. J. Hyman, M. C. Holeva, N. R. Thomson, S. D. Bentley, L. J. C. Churcher, K. Mungall, R. Atkin, N. Bason, K. Brooks, T. Chillingworth, K. Clark, J. Doggett, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, H. Norbertczak, D. Ormond, C. Price, M. A. Quail, M. Sanders, D. Walker, S. Whitehead, G. P. C. Salmond, P. R. J. Birch, J. Parkhill, and I. K. Toth. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 101:11105-11110. - PMC - PubMed
-
- Burkholder, W. H., L. A. McFadden, and A. W. Dimock. 1953. A bacterial blight of chrysanthemums. Phytopathology 43:522-526.
-
- De Boer, S. H., and M. Sasser. 1986. Differentiation of Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica on the basis of cellular fatty acid composition. Can. J. Microbiol. 32:796-800.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

