Common variations in BARD1 influence susceptibility to high-risk neuroblastoma
- PMID: 19412175
- PMCID: PMC2753610
- DOI: 10.1038/ng.374
Common variations in BARD1 influence susceptibility to high-risk neuroblastoma
Abstract
We conducted a SNP-based genome-wide association study (GWAS) focused on the high-risk subset of neuroblastoma. As our previous unbiased GWAS showed strong association of common 6p22 SNP alleles with aggressive neuroblastoma, we restricted our analysis here to 397 high-risk cases compared to 2,043 controls. We detected new significant association of six SNPs at 2q35 within the BARD1 locus (P(allelic) = 2.35 x 10(-9)-2.25 x 10(-8)). We confirmed each SNP association in a second series of 189 high-risk cases and 1,178 controls (P(allelic) = 7.90 x 10(-7)-2.77 x 10(-4)). We also tested the two most significant SNPs (rs6435862, rs3768716) in two additional independent high-risk neuroblastoma case series, yielding combined allelic odds ratios of 1.68 each (P = 8.65 x 10(-18) and 2.74 x 10(-16), respectively). We also found significant association with known BARD1 nonsynonymous SNPs. These data show that common variation in BARD1 contributes to the etiology of the aggressive and most clinically relevant subset of human neuroblastoma.
Figures
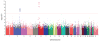

References
-
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. - PubMed
-
- Hero B, et al. Localized infant neuroblastomas often show spontaneous regression: results of the prospective trials NB95-S and NB97. J Clin Oncol. 2008;26:1504–10. - PubMed
-
- Nickerson HJ, et al. Favorable biology and outcome of stage IV-S neuroblastoma with supportive care or minimal therapy: a Children’s Cancer Group study. J Clin Oncol. 2000;18:477–86. - PubMed
-
- Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. New England Journal of Medicine. 1999;341:1165–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

