Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs
- PMID: 19412185
- PMCID: PMC2765218
- DOI: 10.1038/nbt.1539
Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs
Abstract
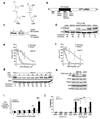
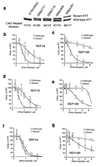
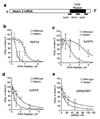
Expanded trinucleotide repeats cause many neurological diseases. These include Machado-Joseph disease (MJD) and Huntington's disease (HD), which are caused by expanded CAG repeats within an allele of the ataxin-3 (ATXN3) and huntingtin (HTT) genes, respectively. Silencing expression of these genes is a promising therapeutic strategy, but indiscriminate inhibition of both the mutant and wild-type alleles may lead to toxicity, and allele-specific approaches have required polymorphisms that differ among individuals. We report that peptide nucleic acid and locked nucleic acid antisense oligomers that target CAG repeats can preferentially inhibit mutant ataxin-3 and HTT protein expression in cultured cells. Duplex RNAs were less selective than single-stranded oligomers. The activity of the peptide nucleic acids does not involve inhibition of transcription, and differences in mRNA secondary structure or the number of oligomer binding sites may be important. Antisense oligomers that discriminate between wild-type and mutant genes on the basis of repeat length may offer new options for developing treatments for MJD, HD and related hereditary diseases.
Figures

Comment in
-
Expanded CAG repeats in the crosshairs.Nat Biotechnol. 2009 May;27(5):451-2. doi: 10.1038/nbt0509-451. Nat Biotechnol. 2009. PMID: 19430450 No abstract available.
References
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. - PubMed
-
- Paulson HL. Dominantly inherited ataxias: Lessons learned from Machado-Joseph disease/spinocerbeller ataxia type 3. Semin. Neurol. 2007;27:133–142. - PubMed
-
- Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. - PubMed
-
- Gusella JF, MacDonald ME. Huntington’s disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci. 2006;31:533–540. - PubMed
-
- Kremer B, et al. A worldwide study of the Huntington’s disease mutation: The sensitivity and specificity of measuring CAG repeats. New Engl. J. Med. 1994;330:1401–1406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

