A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation
- PMID: 19412532
- PMCID: PMC2671163
- DOI: 10.1371/journal.pcbi.1000382
A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation
Abstract
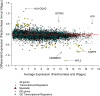
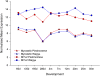
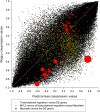
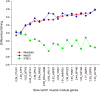
Transcription factor (TF) regulation is often post-translational. TF modifications such as reversible phosphorylation and missense mutations, which can act independent of TF expression level, are overlooked by differential expression analysis. Using bovine Piedmontese myostatin mutants as proof-of-concept, we propose a new algorithm that correctly identifies the gene containing the causal mutation from microarray data alone. The myostatin mutation releases the brakes on Piedmontese muscle growth by translating a dysfunctional protein. Compared to a less muscular non-mutant breed we find that myostatin is not differentially expressed at any of ten developmental time points. Despite this challenge, the algorithm identifies the myostatin 'smoking gun' through a coordinated, simultaneous, weighted integration of three sources of microarray information: transcript abundance, differential expression, and differential wiring. By asking the novel question "which regulator is cumulatively most differentially wired to the abundant most differentially expressed genes?" it yields the correct answer, "myostatin". Our new approach identifies causal regulatory changes by globally contrasting co-expression network dynamics. The entirely data-driven 'weighting' procedure emphasises regulatory movement relative to the phenotypically relevant part of the network. In contrast to other published methods that compare co-expression networks, significance testing is not used to eliminate connections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

