Living-cell microarrays
- PMID: 19413510
- PMCID: PMC3208259
- DOI: 10.1146/annurev.bioeng.10.061807.160502
Living-cell microarrays
Abstract
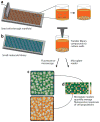
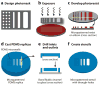
Living cells are remarkably complex. To unravel this complexity, living-cell assays have been developed that allow delivery of experimental stimuli and measurement of the resulting cellular responses. High-throughput adaptations of these assays, known as living-cell microarrays, which are based on microtiter plates, high-density spotting, microfabrication, and microfluidics technologies, are being developed for two general applications: (a) to screen large-scale chemical and genomic libraries and (b) to systematically investigate the local cellular microenvironment. These emerging experimental platforms offer exciting opportunities to rapidly identify genetic determinants of disease, to discover modulators of cellular function, and to probe the complex and dynamic relationships between cells and their local environment.
Figures





References
-
- Ingber DE. Mechanical control of tissue morphogenesis during embryological development. Int J Dev Biol. 2006;50:255–66. - PubMed
-
- Werner S, Krieg T, Smola H. Keratinocyte-fibroblast interactions in wound healing. J Investig Dermatol. 2007;127:998–1008. - PubMed
-
- Saez JC, Berthoud VM, Branes MC, Martinez AD, Beyer EC. Plasma membrane channels formed by connexins: their regulation and functions. Physiol Rev. 2003;83:1359–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

