A functional analysis of the pyrimidine catabolic pathway in Arabidopsis
- PMID: 19413687
- PMCID: PMC2713857
- DOI: 10.1111/j.1469-8137.2009.02843.x
A functional analysis of the pyrimidine catabolic pathway in Arabidopsis
Abstract
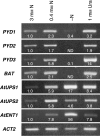
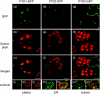
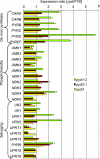
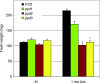
* Reductive catabolism of pyrimidine nucleotides occurs via a three-step pathway in which uracil is degraded to beta-alanine, CO(2) and NH(3) through sequential activities of dihydropyrimidine dehydrogenase (EC 1.3.1.2, PYD1), dihydropyrimidinase (EC 3.5.2.2, PYD2) and beta-ureidopropionase (EC 3.5.1.6, PYD3). * A proposed function of this pathway, in addition to the maintenance of pyrimidine homeostasis, is the recycling of pyrimidine nitrogen to general nitrogen metabolism. PYD expression and catabolism of [2-(14)C]-uracil are markedly elevated in response to nitrogen limitation in plants, which can utilize uracil as a nitrogen source. * PYD1, PYD2 and PYD3 knockout mutants were used for functional analysis of this pathway in Arabidopsis. pyd mutants exhibited no obvious phenotype under optimal growing conditions. pyd2 and pyd3 mutants were unable to catabolize [2-(14)C]-uracil or to grow on uracil as the sole nitrogen source. By contrast, catabolism of uracil was reduced by only 40% in pyd1 mutants, and pyd1 seedlings grew nearly as well as wild-type seedlings with a uracil nitrogen source. These results confirm PYD1 function and suggest the possible existence of another, as yet unknown, activity for uracil degradation to dihydrouracil in this plant. * The localization of PYD-green fluorescent protein fusions in the plastid (PYD1), secretory system (PYD2) and cytosol (PYD3) suggests potentially complex metabolic regulation.
Figures




References
-
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42.
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
-
- Andersen B, Lundgren S, Dobritzsch D, Piškur J. A recruited protease is involved in catabolism of pyrimidines. Journal of Molecular Biology. 2008a;379:243–250. - PubMed
-
- Andersen G, Björnberg O, Polakova S, Pynyaha Y, Rasmussen A, Møller K, Hofer A, Moritz T, Sandrini MPB, Merico A-M, et al. A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. Journal of Molecular Biology. 2008b;380:656–666. - PubMed
-
- Ashihara H. Pattern of uracil metabolism during ageing of tobacco leaves. Acta Physiologiae Plantarum. 1981;3:77–84.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

