Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis
- PMID: 19413989
- PMCID: PMC3297759
- DOI: 10.1016/j.bpj.2008.11.075
Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis
Abstract
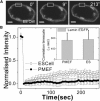
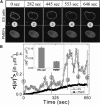
Cellular differentiation and developmental programs require changing patterns of gene expression. Recent experiments have revealed that chromatin organization is highly dynamic within living cells, suggesting possible mechanisms to alter gene expression programs, yet the physical basis of this organization is unclear. In this article, we contrast the differences in the dynamic organization of nuclear architecture between undifferentiated mouse embryonic stem cells and terminally differentiated primary mouse embryonic fibroblasts. Live-cell confocal tracking of nuclear lamina evidences highly flexible nuclear architecture within embryonic stem cells as compared to primary mouse embryonic fibroblasts. These cells also exhibit significant changes in histone and heterochromatin binding proteins correlated with their distinct epigenetic signatures as quantified by immunofluorescence analysis. Further, we follow histone dynamics during the development of the Drosophila melanogaster embryo, which gives an insight into spatio-temporal evolution of chromatin plasticity in an organismal context. Core histone dynamics visualized by fluorescence recovery after photobleaching, fluorescence correlation spectroscopy, and fluorescence anisotropy within the developing embryo, revealed an intriguing transition from plastic to frozen chromatin assembly synchronous with cellular differentiation. In the embryo, core histone proteins are highly mobile before cellularization, actively exchanging with the pool in the yolk. This hyperdynamic mobility decreases as cellularization and differentiation programs set in. These findings reveal a direct correlation between the dynamic transitions in chromatin assembly with the onset of cellular differentiation and developmental programs.
Figures




References
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Lanctot C., Cheutin T., Cremer M., Cavalli G., Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. - PubMed
-
- Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. - PubMed
-
- Luger K., Hansen J.C. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15:188–196. - PubMed
-
- Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

