Subject-specific, multiscale simulation of electrophysiology: a software pipeline for image-based models and application examples
- PMID: 19414456
- PMCID: PMC2696107
- DOI: 10.1098/rsta.2008.0314
Subject-specific, multiscale simulation of electrophysiology: a software pipeline for image-based models and application examples
Abstract
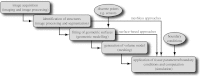
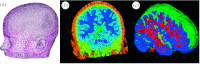
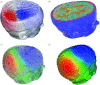
Many simulation studies in biomedicine are based on a similar sequence of processing steps, starting from images and running through geometric model generation, assignment of tissue properties, numerical simulation and visualization of the results--a process known as image-based geometric modelling and simulation. We present an overview of software systems for implementing such a sequence both within highly integrated problem-solving environments and in the form of loosely integrated pipelines. Loose integration in this case indicates that individual programs function largely independently but communicate through files of a common format and support simple scripting, so as to automate multiple executions wherever possible. We then describe three specific applications of such pipelines to translational biomedical research in electrophysiology.
Figures
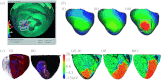
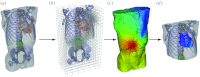
References
-
- Callahan, M., Cole, M., Shepherd, J., Stinstra, J. & Johnson, C. 2009 A meshing pipeline for biomedical computing. Eng. Comput 25, 115–130. (10.1007/s00366-008-0106-1) - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
