A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation
- PMID: 19414554
- PMCID: PMC2715034
- DOI: 10.1084/jem.20081621
A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation
Abstract
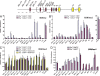
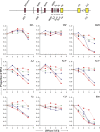
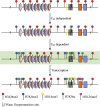
A tissue-specific transcriptional enhancer, Emu, has been implicated in developmentally regulated recombination and transcription of the immunoglobulin heavy chain (IgH) gene locus. We demonstrate that deleting 220 nucleotides that constitute the core Emu results in partially active locus, characterized by reduced histone acetylation, chromatin remodeling, transcription, and recombination, whereas other hallmarks of tissue-specific locus activation, such as loss of H3K9 dimethylation or gain of H3K4 dimethylation, are less affected. These observations define Emu-independent and Emu-dependent phases of locus activation that reveal an unappreciated epigenetic hierarchy in tissue-specific gene expression.
Figures


References
-
- Bulger M., Sawado T., Schubeler D., Groudine M. 2002. ChIPs of the beta-globin locus: unraveling gene regulation within an active domain.Curr. Opin. Genet. Dev. 12:170–177 - PubMed
-
- Felsenfeld G., Burgess-Beusse B., Farrell C., Gaszner M., Ghirlando R., Huang S., Jin C., Litt M., Magdinier F., Mutskov V., et al. 2004. Chromatin boundaries and chromatin domains.Cold Spring Harb. Symp. Quant. Biol. 69:245–250 - PubMed
-
- Bender M.A., Bulger M., Close J., Groudine M. 2000. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region.Mol. Cell. 5:387–393 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

