Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus
- PMID: 19414564
- PMCID: PMC2710164
- DOI: 10.1534/genetics.109.101642
Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus
Abstract
Yield is the most important and complex trait for the genetic improvement of crops. Although much research into the genetic basis of yield and yield-associated traits has been reported, in each such experiment the genetic architecture and determinants of yield have remained ambiguous. One of the most intractable problems is the interaction between genes and the environment. We identified 85 quantitative trait loci (QTL) for seed yield along with 785 QTL for eight yield-associated traits, from 10 natural environments and two related populations of rapeseed. A trait-by-trait meta-analysis revealed 401 consensus QTL, of which 82.5% were clustered and integrated into 111 pleiotropic unique QTL by meta-analysis, 47 of which were relevant for seed yield. The complexity of the genetic architecture of yield was demonstrated, illustrating the pleiotropy, synthesis, variability, and plasticity of yield QTL. The idea of estimating indicator QTL for yield QTL and identifying potential candidate genes for yield provides an advance in methodology for complex traits.
Figures

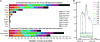
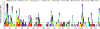

References
-
- Arcade, A., A. Labourdette, M. Falque, B. Mangin, F. Chardon et al., 2004. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20 2324–2326. - PubMed
-
- Ashikari, M., H. Sakakibara, S. Lin, T. Yamamoto, T. Takashi et al., 2005. Cytokinin oxidase regulates rice grain production. Science 309 741–745. - PubMed
-
- Brown, J. K., 2002. Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 5 339–344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

