Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae
- PMID: 19414575
- PMCID: PMC2704628
- DOI: 10.1128/AAC.00880-08
Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae
Abstract
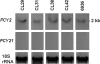
The aim of this work was to elucidate the molecular mechanisms of flucytosine (5FC) resistance and 5FC/fluconazole (FLC) cross-resistance in 11 genetically and epidemiologically unrelated clinical isolates of Candida lusitaniae. We first showed that the levels of transcription of the FCY2 gene encoding purine-cytosine permease (PCP) in the isolates were similar to that in the wild-type strain, 6936. Nucleotide sequencing of the FCY2 alleles revealed that 5FC and 5FC/FLC resistance could be correlated with a cytosine-to-thymine substitution at nucleotide 505 in the fcy2 genes of seven clinical isolates, resulting in a nonsense mutation and in a putative nonfunctional truncated PCP of 168 amino acids. Reintroducing a FCY2 wild-type allele at the fcy2 locus of a ura3 auxotrophic strain derived from the clinical isolate CL38 fcy2(C505T) restored levels of susceptibility to antifungals comparable to those of the wild-type strains. In the remaining four isolates, a polymorphic nucleotide was found in FCY1 where the nucleotide substitution T26C resulted in the amino acid replacement M9T in cytosine deaminase. Introducing this mutated allele into a 5FC- and 5FC/FLC-resistant fcy1Delta strain failed to restore antifungal susceptibility, while susceptibility was obtained by introducing a wild-type FCY1 allele. We thus found a correlation between the fcy1 T26C mutation and both 5FC and 5FC/FLC resistances. We demonstrated that only two genetic events occurred in 11 unrelated clinical isolates of C. lusitaniae to support 5FC and 5FC/FLC resistance: either the nonsense mutation C505T in the fcy2 gene or the missense mutation T26C in the fcy1 gene.
Figures

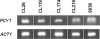

Similar articles
-
Inactivation of the FCY2 gene encoding purine-cytosine permease promotes cross-resistance to flucytosine and fluconazole in Candida lusitaniae.Antimicrob Agents Chemother. 2005 Aug;49(8):3101-8. doi: 10.1128/AAC.49.8.3101-3108.2005. Antimicrob Agents Chemother. 2005. PMID: 16048910 Free PMC article.
-
Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance.Antimicrob Agents Chemother. 2007 Jan;51(1):369-71. doi: 10.1128/AAC.00824-06. Epub 2006 Oct 23. Antimicrob Agents Chemother. 2007. PMID: 17060521 Free PMC article.
-
Mutational analysis of flucytosine resistance in Candida glabrata.Antimicrob Agents Chemother. 2010 Nov;54(11):4733-8. doi: 10.1128/AAC.00605-10. Epub 2010 Sep 7. Antimicrob Agents Chemother. 2010. PMID: 20823283 Free PMC article.
-
The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans.Crit Rev Microbiol. 1987;15(1):45-56. doi: 10.3109/10408418709104447. Crit Rev Microbiol. 1987. PMID: 3319420 Review.
-
Antifungal drug resistance in pathogenic fungi.Med Mycol. 1998;36 Suppl 1:119-28. Med Mycol. 1998. PMID: 9988500 Review.
Cited by
-
Comparative Genomics for the Elucidation of Multidrug Resistance in Candida lusitaniae.mBio. 2019 Dec 24;10(6):e02512-19. doi: 10.1128/mBio.02512-19. mBio. 2019. PMID: 31874914 Free PMC article.
-
Molecular Mechanisms Associated with Antifungal Resistance in Pathogenic Candida Species.Cells. 2023 Nov 19;12(22):2655. doi: 10.3390/cells12222655. Cells. 2023. PMID: 37998390 Free PMC article. Review.
-
Striking Back against Fungal Infections: The Utilization of Nanosystems for Antifungal Strategies.Int J Mol Sci. 2021 Sep 18;22(18):10104. doi: 10.3390/ijms221810104. Int J Mol Sci. 2021. PMID: 34576268 Free PMC article. Review.
-
5-fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in Cryptococcus.Nat Commun. 2020 Jan 8;11(1):127. doi: 10.1038/s41467-019-13890-z. Nat Commun. 2020. PMID: 31913284 Free PMC article.
-
The Cryptococcus neoformans transcriptome at the site of human meningitis.mBio. 2014 Feb 4;5(1):e01087-13. doi: 10.1128/mBio.01087-13. mBio. 2014. PMID: 24496797 Free PMC article.
References
-
- Al Mosaid, A., D. J. Sullivan, I. Polacheck, F. A. Shaheen, O. Soliman, S. Al Hedaithy, S. Al Thawad, M. Kabadaya, and D. C. Coleman. 2005. Novel 5-flucytosine-resistant clade of Candida dubliniensis from Saudi Arabia and Egypt identified by Cd25 fingerprinting. J. Clin. Microbiol. 43:4026-4036. - PMC - PubMed
-
- Balkis, M. M., S. D. Leidich, P. K. Mukherjee, and M. A. Ghannoum. 2002. Mechanisms of fungal resistance: an overview. Drugs 62:1025-1040. - PubMed
-
- Barker, K. S., and P. D. Rogers. 2006. Recent insights into the mechanisms of antifungal resistance. Curr. Infect. Dis. Rep. 8:449-456. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

